| Biological Process |
| dendritic spine morphogenesis |
| protein kinase B signaling cascade |
| negative regulation of phosphatidylinositol 3-kinase cascade |
| negative regulation of cell proliferation |
| dentate gyrus development |
| heart development |
| negative regulation of ribosome biogenesis |
| negative regulation of cell aging |
| cell migration |
| social behavior |
| negative regulation of synaptic vesicle clustering |
| epidermal growth factor receptor signaling pathway |
| forebrain morphogenesis |
| endothelial cell migration |
| neuron-neuron synaptic transmission |
| fibroblast growth factor receptor signaling pathway |
| locomotor rhythm |
| T cell receptor signaling pathway |
| positive regulation of excitatory postsynaptic membrane potential |
| nerve growth factor receptor signaling pathway |
| long-term synaptic potentiation |
| angiogenesis |
| learning or memory |
| positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process |
| male mating behavior |
| inositol phosphate dephosphorylation |
| negative regulation of organ growth |
| postsynaptic density assembly |
| cell proliferation |
| positive regulation of sequence-specific DNA binding transcription factor activity |
| phosphatidylinositol biosynthetic process |
| prepulse inhibition |
| negative regulation of axonogenesis |
| induction of apoptosis |
| apoptotic process |
| presynaptic membrane assembly |
| regulation of B cell apoptotic process |
| negative regulation of cell size |
| negative regulation of G1/S transition of mitotic cell cycle |
| positive regulation of cell proliferation |
| prostate gland growth |
| negative regulation of cyclin-dependent protein kinase activity involved in G1/S |
| canonical Wnt receptor signaling pathway |
| negative regulation of apoptotic process |
| regulation of myeloid cell apoptotic process |
| negative regulation of dendritic spine morphogenesis |
| cardiac muscle tissue development |
| negative regulation of protein kinase B signaling cascade |
| rhythmic synaptic transmission |
| negative regulation of epithelial cell proliferation |
| negative regulation of excitatory postsynaptic membrane potential |
| activation of mitotic anaphase-promoting complex activity |
| synapse maturation |
| negative regulation of focal adhesion assembly |
| negative regulation of cell migration |
| central nervous system myelin maintenance |
| phosphatidylinositol dephosphorylation |
| negative regulation of myelination |
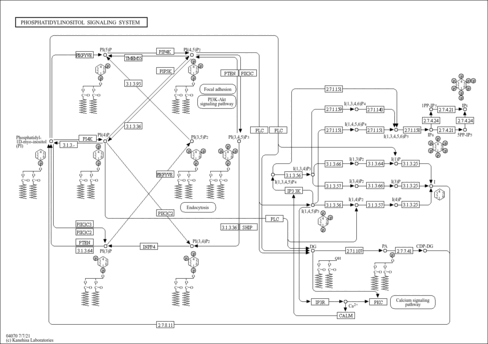
| phosphatidylinositol-mediated signaling |
| central nervous system neuron axonogenesis |
| maternal behavior |
| protein stabilization |
| multicellular organismal response to stress |
| Cellular Component |
| cytosol |
| myelin sheath adaxonal region |
| Schmidt-Lanterman incisure |
| nucleus |
| internal side of plasma membrane |
| PML body |
| neuron projection |
| Function |
| metal ion binding |
| protein tyrosine/serine/threonine phosphatase activity |
| hydrolase activity, acting on ester bonds |
| binding |
| catalytic activity |
| hydrolase activity |
| phosphoric ester hydrolase activity |
| phosphatase activity |
| magnesium ion binding |
| protein binding |
| inositol or phosphatidylinositol phosphatase activity |
| phosphatidylinositol bisphosphate phosphatase activity |
| phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity |
| phosphoprotein phosphatase activity |
| protein domain specific binding |
| protein tyrosine phosphatase activity |
| pdz domain binding |
| ion binding |
| phosphatidylinositol trisphosphate phosphatase activity |
| cation binding |
| phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity |
| Molecular Function |
| protein tyrosine/serine/threonine phosphatase activity |
| protein serine/threonine phosphatase activity |
| magnesium ion binding |
| phosphatidylinositol-3-phosphatase activity |
| inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity |
| phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity |
| phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity |
| lipid binding |
| protein tyrosine phosphatase activity |
| Process |
| regulation of cell-substrate adhesion |
| posttranscriptional regulation of gene expression |
| inositol metabolic process |
| regulation of cell-matrix adhesion |
| biological regulation |
| glycerophospholipid metabolic process |
| negative regulation of cell-matrix adhesion |
| regulation of biological process |
| phosphoinositide metabolic process |
| negative regulation of focal adhesion assembly |
| regulation of metabolic process |
| phosphoinositide dephosphorylation |
| inositol phosphate dephosphorylation |
| regulation of macromolecule metabolic process |
| regulation of protein stability |
| regulation of gene expression |
| regulation of locomotion |
| cellular metabolic process |
| regulation of cell motility |
| small molecule metabolic process |
| regulation of cell migration |
| alcohol metabolic process |
| phosphorus metabolic process |
| negative regulation of cell migration |
| regulation of cellular process |
| phosphate metabolic process |
| negative regulation of cell proliferation |
| negative regulation of signal transduction |
| dephosphorylation |
| regulation of cell communication |
| negative regulation of intracellular protein kinase cascade |
| protein amino acid dephosphorylation |
| regulation of signal transduction |
| negative regulation of protein kinase b signaling cascade |
| metabolic process |
| organophosphate metabolic process |
| polyol metabolic process |
| regulation of cell adhesion |
| regulation of cell proliferation |
| phospholipid metabolic process |