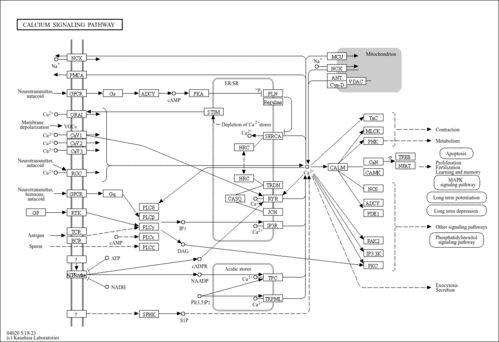
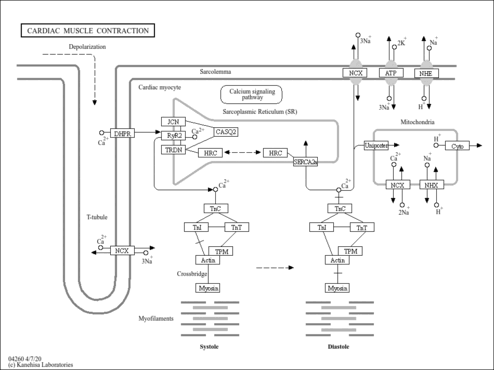
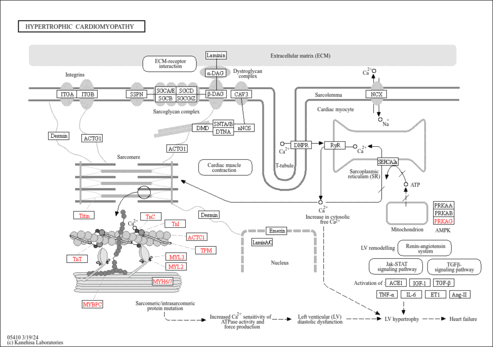
Showing Protein Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (CDBP01365)
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HMDB Protein ID | CDBP01365 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Name | ATP2A2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Type | Enzyme | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Function | Involved in ATP binding | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Specific Function | This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the translocation of calcium from the cytosol to the sarcoplasmic reticulum lumen. Isoform 2 is involved in the regulation of the contraction/relaxation cycle. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO Classification |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Location |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chromosome Location | 12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Locus | 12q24.11 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SNPs | ATP2A2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Sequence |
>3129 bp ATGGAGAACGCGCACACCAAGACGGTGGAGGAGGTGCTGGGCCACTTCGGCGTCAACGAG AGTACGGGGCTGAGCCTGGAACAGGTCAAGAAGCTTAAGGAGAGATGGGGCTCCAACGAG TTACCGGCTGAAGAAGGAAAAACCTTGCTGGAACTTGTGATTGAGCAGTTTGAAGACTTG CTAGTTAGGATTTTATTACTGGCAGCATGTATATCTTTTGTTTTGGCTTGGTTTGAAGAA GGTGAAGAAACAATTACAGCCTTTGTAGAACCTTTTGTAATTTTACTCATATTAGTAGCC AATGCAATTGTGGGTGTATGGCAGGAAAGAAATGCTGAAAATGCCATCGAAGCCCTTAAG GAATATGAGCCTGAAATGGGCAAAGTGTATCGACAGGACAGAAAGAGTGTGCAGCGGATT AAAGCTAAAGACATAGTTCCTGGTGATATTGTAGAAATTGCTGTTGGTGACAAAGTTCCT GCTGATATAAGGTTAACTTCCATCAAATCTACCACACTAAGAGTTGACCAGTCAATTCTC ACAGGTGAATCTGTCTCTGTCATCAAGCACACTGATCCCGTCCCTGACCCACGAGCTGTC AACCAAGATAAAAAGAACATGCTGTTTTCTGGTACAAACATTGCTGCTGGGAAAGCTATG GGAGTGGTGGTAGCAACTGGAGTTAACACCGAAATTGGCAAGATCCGGGATGAAATGGTG GCAACAGAACAGGAGAGAACACCCCTTCAGCAAAAACTAGATGAATTTGGGGAACAGCTT TCCAAAGTCATCTCCCTTATTTGCATTGCAGTCTGGATCATAAATATTGGGCACTTCAAT GACCCGGTTCATGGAGGGTCCTGGATCAGAGGTGCTATTTACTACTTTAAAATTGCAGTG GCCCTGGCTGTAGCAGCCATTCCTGAAGGTCTGCCTGCAGTCATCACCACCTGCCTGGCT CTTGGAACTCGCAGAATGGCAAAGAAAAATGCCATTGTTCGAAGCCTCCCGTCTGTGGAA ACCCTTGGTTGTACTTCTGTTATCTGCTCAGACAAGACTGGTACACTTACAACAAACCAG ATGTCAGTCTGCAGGATGTTCATTCTGGACAGAGTGGAAGGTGATACTTGTTCCCTTAAT GAGTTTACCATAACTGGATCAACTTATGCACCTATTGGAGAAGTGCATAAAGATGATAAA CCAGTGAATTGTCACCAGTATGATGGTCTGGTAGAATTAGCAACAATTTGTGCTCTTTGT AATGACTCTGCTTTGGATTACAATGAGGCAAAGGGTGTGTATGAAAAAGTTGGAGAAGCT ACAGAGACTGCTCTCACTTGCCTAGTAGAGAAGATGAATGTATTTGATACCGAATTGAAG GGTCTTTCTAAAATAGAACGTGCAAATGCCTGCAACTCAGTCATTAAACAGCTGATGAAA AAGGAATTCACTCTAGAGTTTTCACGTGACAGAAAGTCAATGTCGGTTTACTGTACACCA AATAAACCAAGCAGGACATCAATGAGCAAGATGTTTGTGAAGGGTGCTCCTGAAGGTGTC ATTGACAGGTGCACCCACATTCGAGTTGGAAGTACTAAGGTTCCTATGACCTCTGGAGTC AAACAGAAGATCATGTCTGTCATTCGAGAGTGGGGTAGTGGCAGCGACACACTGCGATGC CTGGCCCTGGCCACTCATGACAACCCACTGAGAAGAGAAGAAATGCACCTTGAGGACTCT GCCAACTTTATTAAATATGAGACCAATCTGACCTTCGTTGGCTGCGTGGGCATGCTGGAT CCTCCGAGAATCGAGGTGGCCTCCTCCGTGAAGCTGTGCCGGCAAGCAGGCATCCGGGTC ATCATGATCACTGGGGACAACAAGGGCACTGCTGTGGCCATCTGTCGCCGCATCGGCATC TTCGGGCAGGATGAGGACGTGACGTCAAAAGCTTTCACAGGCCGGGAGTTTGATGAACTC AACCCCTCCGCCCAGCGAGACGCCTGCCTGAACGCCCGCTGTTTTGCTCGAGTTGAACCC TCCCACAAGTCTAAAATCGTAGAATTTCTTCAGTCTTTTGATGAGATTACAGCTATGACT GGCGATGGCGTGAACGATGCTCCTGCTCTGAAGAAAGCCGAGATTGGCATTGCTATGGGC TCTGGCACTGCGGTGGCTAAAACCGCCTCTGAGATGGTCCTGGCGGATGACAACTTCTCC ACCATTGTGGCTGCCGTTGAGGAGGGGCGGGCAATCTACAACAACATGAAACAGTTCATC CGCTACCTCATCTCGTCCAACGTCGGGGAAGTTGTCTGTATTTTCCTGACAGCAGCCCTT GGATTTCCCGAGGCTTTGATTCCTGTTCAGCTGCTCTGGGTCAATCTGGTGACAGATGGC CTGCCTGCCACTGCACTGGGGTTCAACCCTCCTGATCTGGACATCATGAATAAACCTCCC CGGAACCCAAAGGAACCATTGATCAGCGGGTGGCTCTTTTTCCGTTACTTGGCTATTGGC TGTTACGTCGGCGCTGCTACCGTGGGTGCTGCTGCATGGTGGTTCATTGCTGCTGACGGT GGTCCAAGAGTGTCCTTCTACCAGCTGAGTCATTTCCTACAGTGTAAAGAGGACAACCCG GACTTTGAAGGCGTGGATTGTGCAATCTTTGAATCCCCATACCCGATGACAATGGCGCTC TCTGTTCTAGTAACTATAGAAATGTGTAACGCCCTCAACAGCTTGTCCGAAAACCAGTCC TTGCTGAGGATGCCCCCCTGGGAGAACATCTGGCTCGTGGGCTCCATCTGCCTGTCCATG TCACTCCACTTCCTGATCCTCTATGTCGAACCCTTGCCACTCATCTTCCAGATCACACCG CTGAACGTGACCCAGTGGCTGATGGTGCTGAAAATCTCCTTGCCCGTGATTCTCATGGAT GAGACGCTCAAGTTTGTGGCCCGCAACTACCTGGAACCTGGTAAAGAGTGTGTGCAGCCT GCCACCAAATCCTGCTCGTTCTCGGCATGCACCGATGGGATTTCCTGGCCGTTTGTGCTG CTCATAATGCCCCTGGTGATCTGGGTCTATAGCACAGACACTAACTTTAGCGATATGTTC TGGTCTTGA |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of Residues | 1042 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Weight | 109689.97 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Theoretical pI | 5.366 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pfam Domain Function | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Signals | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transmembrane Regions | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Sequence |
>Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 MENAHTKTVEEVLGHFGVNESTGLSLEQVKKLKERWGSNELPAEEGKTLLELVIEQFEDL LVRILLLAACISFVLAWFEEGEETITAFVEPFVILLILVANAIVGVWQERNAENAIEALK EYEPEMGKVYRQDRKSVQRIKAKDIVPGDIVEIAVGDKVPADIRLTSIKSTTLRVDQSIL TGESVSVIKHTDPVPDPRAVNQDKKNMLFSGTNIAAGKAMGVVVATGVNTEIGKIRDEMV ATEQERTPLQQKLDEFGEQLSKVISLICIAVWIINIGHFNDPVHGGSWIRGAIYYFKIAV ALAVAAIPEGLPAVITTCLALGTRRMAKKNAIVRSLPSVETLGCTSVICSDKTGTLTTNQ MSVCRMFILDRVEGDTCSLNEFTITGSTYAPIGEVHKDDKPVNCHQYDGLVELATICALC NDSALDYNEAKGVYEKVGEATETALTCLVEKMNVFDTELKGLSKIERANACNSVIKQLMK KEFTLEFSRDRKSMSVYCTPNKPSRTSMSKMFVKGAPEGVIDRCTHIRVGSTKVPMTSGV KQKIMSVIREWGSGSDTLRCLALATHDNPLRREEMHLEDSANFIKYETNLTFVGCVGMLD PPRIEVASSVKLCRQAGIRVIMITGDNKGTAVAICRRIGIFGQDEDVTSKAFTGREFDEL NPSAQRDACLNARCFARVEPSHKSKIVEFLQSFDEITAMTGDGVNDAPALKKAEIGIAMG SGTAVAKTASEMVLADDNFSTIVAAVEEGRAIYNNMKQFIRYLISSNVGEVVCIFLTAAL GFPEALIPVQLLWVNLVTDGLPATALGFNPPDLDIMNKPPRNPKEPLISGWLFFRYLAIG CYVGAATVGAAAWWFIAADGGPRVSFYQLSHFLQCKEDNPDFEGVDCAIFESPYPMTMAL SVLVTIEMCNALNSLSENQSLLRMPPWENIWLVGSICLSMSLHFLILYVEPLPLIFQITP LNVTQWLMVLKISLPVILMDETLKFVARNYLEPGKECVQPATKSCSFSACTDGISWPFVL LIMPLVIWVYSTDTNFSDMFWS |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GenBank ID Protein | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProtKB/Swiss-Prot ID | P16615 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProtKB/Swiss-Prot Entry Name | AT2A2_HUMAN | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB IDs | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GenBank Gene ID | M23114 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GeneCard ID | ATP2A2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GenAtlas ID | ATP2A2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HGNC ID | HGNC:812 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||