| Identification |
|---|
| HMDB Protein ID
| CDBP01325 |
| Secondary Accession Numbers
| Not Available |
| Name
| Ribosomal protein S6 kinase beta-1 |
| Description
| Not Available |
| Synonyms
|
- 70 kDa ribosomal protein S6 kinase 1
- P70S6K1
- Ribosomal protein S6 kinase I
- S6K-beta-1
- S6K1
- Serine/threonine-protein kinase 14A
- p70 S6 kinase alpha
- p70 S6K-alpha
- p70 S6KA
- p70 ribosomal S6 kinase alpha
- p70-S6K 1
|
| Gene Name
| RPS6KB1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in protein serine/threonine kinase activity |
| Specific Function
| Acts to integrate nutrient and growth factor signals in regulation of protein synthesis, cell proliferation, cell growth, cell cycle progression and cell survival. Downstream effector of the mTOR signaling pathway. Phosphorylates specifically ribosomal protein S6 in response to insulin or several classes of mitogens. During translation initiation, the inactive form associatess with the eIF-3 complex under conditions of nutrient depletion. Mitogenic stimulation leads to phosphorylation and dissociation from the eIF-3 complex and the free activated form can phosphorylate other translational targets including EIF4B. Promotes protein synthesis by phosphorylating PDCD4 at 'Ser-67' and targeting it for degradation. Phosphorylates RICTOR leading to regulation of mammalian target of rapamycin complex 2 (mTORC2) signaling; probably phosphorylates RICTOR at 'Thr-1135'. Phosphorylates IRS1 at multiple serine residues coupled with insulin resistance; probably phosphorylates IRS1 at 'Ser-270'. Required for TNF-alpha induced IRS-1 degradation. Phosphorylates EEF2K in response to IGF1 and inhibits EEF2K activity. Phosphorylates BAD at 'Ser-99' in response to IGF1 leading to BAD inactivation and inhibition of BAD-induced apoptosis. Phosphorylates mitochondrial RMP leading to dissociation of a RMP:PPP1CC complex; probably phosphorylates RMP at 'Ser-99'. The free mitochondrial PPP1CC can dephosphorylate RPS6KB1 at Thr-412 which is proposed to be a negative feed back mechanism for the RPS6KB1 antiapoptotic function. Phosphorylates GSK3B at 'Ser-9' under conditions leading to loss of the TSC1-TSC2 complex. Phosphorylates POLDIP3 |
| GO Classification
|
| Function |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| kinase activity |
| nucleoside binding |
| purine nucleoside binding |
| adenyl nucleotide binding |
| adenyl ribonucleotide binding |
| atp binding |
| protein kinase activity |
| protein serine/threonine kinase activity |
| binding |
| catalytic activity |
| Process |
| regulation of cellular process |
| signal transduction |
| protein amino acid phosphorylation |
| phosphorylation |
| phosphorus metabolic process |
| phosphate metabolic process |
| metabolic process |
| biological regulation |
| regulation of biological process |
| cellular metabolic process |
|
| Cellular Location
|
- Isoform Alpha II:Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Leucine Stimulation on Insulin Signaling | 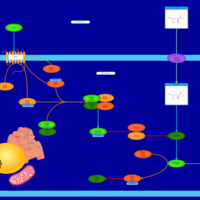  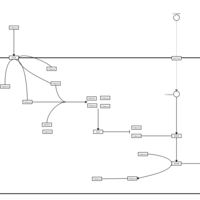 | Not Available | | BCR-ABL Action in CML Pathogenesis | 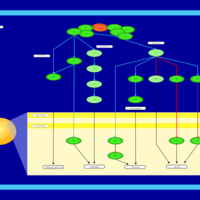 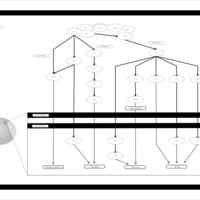 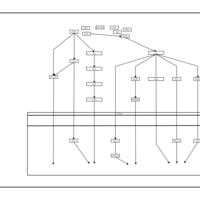 | Not Available | | Imatinib Inhibition of BCR-ABL | 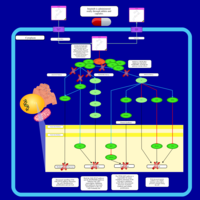 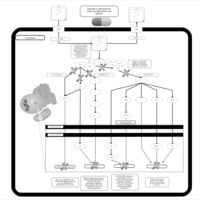  | Not Available | | Dasatinib Inhibition of BCR-ABL | 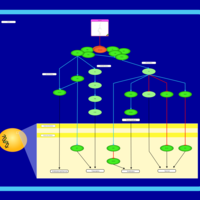 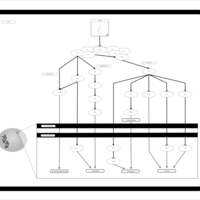 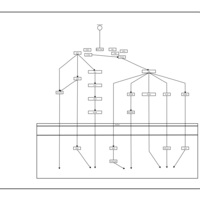 | Not Available | | Nilotinib Inhibition of BCR-ABL | 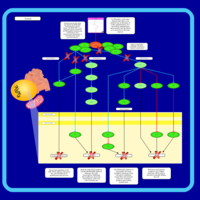 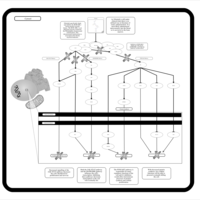  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| Chromosome:1 |
| Locus
| 17q23.1 |
| SNPs
| RPS6KB1 |
| Gene Sequence
|
>1578 bp
ATGAGGCGACGAAGGAGGCGGGACGGCTTTTACCCAGCCCCGGACTTCCGAGACAGGGAA
GCTGAGGACATGGCAGGAGTGTTTGACATAGACCTGGACCAGCCAGAGGACGCGGGCTCT
GAGGATGAGCTGGAGGAGGGGGGTCAGTTAAATGAAAGCATGGACCATGGGGGAGTTGGA
CCATATGAACTTGGCATGGAACATTGTGAGAAATTTGAAATCTCAGAAACTAGTGTGAAC
AGAGGGCCAGAAAAAATCAGACCAGAATGTTTTGAGCTACTTCGGGTACTTGGTAAAGGG
GGCTATGGAAAGGTTTTTCAAGTACGAAAAGTAACAGGAGCAAATACTGGGAAAATATTT
GCCATGAAGGTGCTTAAAAAGGCAATGATAGTAAGAAATGCTAAAGATACAGCTCATACA
AAAGCAGAACGGAATATTCTGGAGGAAGTAAAGCATCCCTTCATCGTGGATTTAATTTAT
GCCTTTCAGACTGGTGGAAAACTCTACCTCATCCTTGAGTATCTCAGTGGAGGAGAACTA
TTTATGCAGTTAGAAAGAGAGGGAATATTTATGGAAGACACTGCCTGCTTTTACTTGGCA
GAAATCTCCATGGCTTTGGGGCATTTACATCAAAAGGGGATCATCTACAGAGACCTGAAG
CCGGAGAATATCATGCTTAATCACCAAGGTCATGTGAAACTAACAGACTTTGGACTATGC
AAAGAATCTATTCATGATGGAACAGTCACACACACATTTTGTGGAACAATAGAATACATG
GCCCCTGAAATCTTGATGAGAAGTGGCCACAATCGTGCTGTGGATTGGTGGAGTTTGGGA
GCATTAATGTATGACATGCTGACTGGAGCACCCCCATTCACTGGGGAGAATAGAAAGAAA
ACAATTGACAAAATCCTCAAATGTAAACTCAATTTGCCTCCCTACCTCACACAAGAAGCC
AGAGATCTGCTTAAAAAGCTGCTGAAAAGAAATGCTGCTTCTCGTCTGGGAGCTGGTCCT
GGGGACGCTGGAGAAGTTCAAGCTCATCCATTCTTTAGACACATTAACTGGGAAGAACTT
CTGGCTCGAAAGGTGGAGCCCCCCTTTAAACCTCTGTTGCAATCTGAAGAGGATGTAAGT
CAGTTTGATTCCAAGTTTACACGTCAGACACCTGTCGACAGCCCAGATGACTCAACTCTC
AGTGAAAGTGCCAATCAGGTCTTTCTGGGTTTTACATATGTGGCTCCATCTGTACTTGAA
AGTGTGAAAGAAAAGTTTTCCTTTGAACCAAAAATCCGATCACCTCGAAGATTTATTGGC
AGCCCACGAACACCTGTCAGCCCAGTCAAATTTTCTCCTGGGGATTTCTGGGGAAGAGGT
GCTTCGGCCAGCACAGCAAATCCTCAGACACCTGTGGAATACCCAATGGAAACAAGTGGC
ATAGAGCAGATGGATGTGACAATGAGTGGGGAAGCATCGGCACCACTTCCAATACGACAG
CCGAACTCTGGGCCATACAAAAAACAAGCTTTTCCCATGATCTCCAAACGGCCAGAGCAC
CTGCGTATGAATCTATGA
|
| Protein Properties |
|---|
| Number of Residues
| 525 |
| Molecular Weight
| 59139.1 |
| Theoretical pI
| 6.64 |
| Pfam Domain Function
|
|
| Signals
|
|
|
Transmembrane Regions
|
|
| Protein Sequence
|
>Ribosomal protein S6 kinase beta-1
MRRRRRRDGFYPAPDFRDREAEDMAGVFDIDLDQPEDAGSEDELEEGGQLNESMDHGGVG
PYELGMEHCEKFEISETSVNRGPEKIRPECFELLRVLGKGGYGKVFQVRKVTGANTGKIF
AMKVLKKAMIVRNAKDTAHTKAERNILEEVKHPFIVDLIYAFQTGGKLYLILEYLSGGEL
FMQLEREGIFMEDTACFYLAEISMALGHLHQKGIIYRDLKPENIMLNHQGHVKLTDFGLC
KESIHDGTVTHTFCGTIEYMAPEILMRSGHNRAVDWWSLGALMYDMLTGAPPFTGENRKK
TIDKILKCKLNLPPYLTQEARDLLKKLLKRNAASRLGAGPGDAGEVQAHPFFRHINWEEL
LARKVEPPFKPLLQSEEDVSQFDSKFTRQTPVDSPDDSTLSESANQVFLGFTYVAPSVLE
SVKEKFSFEPKIRSPRRFIGSPRTPVSPVKFSPGDFWGRGASASTANPQTPVEYPMETSG
IEQMDVTMSGEASAPLPIRQPNSGPYKKQAFPMISKRPEHLRMNL
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| P23443 |
| UniProtKB/Swiss-Prot Entry Name
| KS6B1_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| M60724 |
| GeneCard ID
| RPS6KB1 |
| GenAtlas ID
| RPS6KB1 |
| HGNC ID
| HGNC:10436 |
| References |
|---|
| General References
| Not Available |