Showing Protein Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (CDBP01289)
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HMDB Protein ID | CDBP01289 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Name | ATP2A1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Type | Enzyme | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Function | Involved in ATP binding | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Specific Function | This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the translocation of calcium from the cytosol to the sarcoplasmic reticulum lumen. Contributes to calcium sequestration involved in muscular excitation/contraction. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO Classification |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Location |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
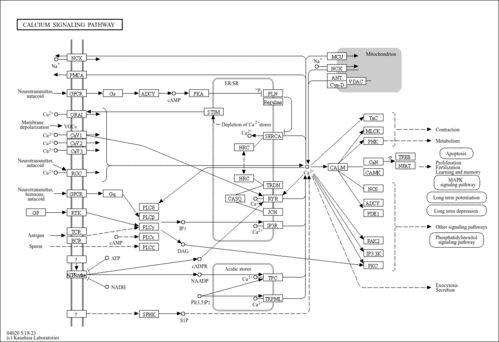
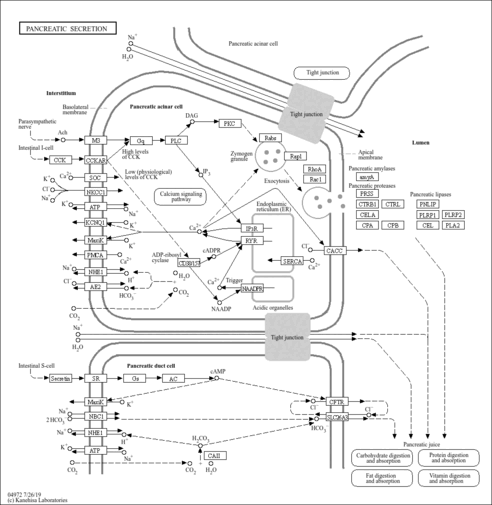
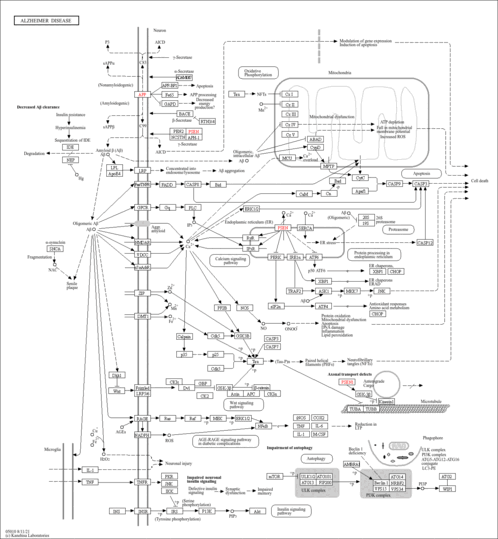
| Pathways |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chromosome Location | 16 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Locus | 16p12.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SNPs | ATP2A1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Sequence |
>3006 bp ATGGAGGCCGCTCATGCTAAAACCACGGAGGAATGTTTGGCCTATTTTGGGGTGAGTGAG ACCACGGGCCTCACCCCGGACCAAGTTAAGCGGAATCTGGAGAAATACGGCCTCAATGAG CTCCCTGCTGAGGAAGGGAAGACCCTGTGGGAGCTGGTGATAGAGCAGTTTGAAGACCTC CTGGTGCGGATTCTCCTCCTGGCCGCATGCATTTCCTTCGTGCTGGCCTGGTTTGAGGAA GGTGAAGAGACCATCACTGCCTTTGTTGAACCCTTTGTCATCCTCTTGATCCTCATTGCC AATGCCATCGTGGGGGTTTGGCAGGAGCGGAACGCAGAGAACGCCATCGAGGCCCTGAAG GAGTATGAGCCAGAGATGGGGAAGGTCTACCGGGCTGACCGCAAGTCAGTGCAAAGGATC AAGGCTCGGGACATCGTCCCTGGGGACATCGTGGAGGTGGCTGTGGGGGACAAAGTCCCT GCAGACATCCGAATCCTCGCCATCAAATCCACCACGCTGCGGGTTGACCAGTCCATCCTG ACAGGCGAGTCTGTATCTGTCATCAAACACACGGAGCCCGTTCCTGACCCCCGAGCTGTC AACCAGGACAAGAAGAACATGCTTTTCTCGGGCACCAACATTGCAGCCGGCAAGGCCTTG GGCATCGTGGCCACCACTGGTGTGGGCACCGAGATTGGGAAGATCCGAGACCAAATGGCT GCCACAGAACAGGACAAGACCCCCTTGCAGCAGAAGCTGGATGAGTTTGGGGAGCAGCTC TCCAAGGTCATCTCCCTCATCTGTGTGGCTGTCTGGCTTATCAACATTGGCCACTTCAAC GACCCCGTCCATGGGGGCTCCTGGTTCCGCGGGGCCATCTACTACTTTAAGATTGCCGTG GCCTTGGCTGTGGCTGCCATCCCCGAAGGTCTTCCTGCAGTCATCACCACCTGCCTGGCC CTGGGTACCCGTCGGATGGCAAAGAAGAATGCCATTGTAAGAAGCTTGCCCTCCGTAGAG ACCCTGGGCTGCACCTCTGTCATCTGTTCCGACAAGACAGGCACCCTCACCACCAACCAG ATGTCTGTCTGCAAGATGTTTATCATTGACAAGGTGGATGGGGACATCTGCCTCCTGAAT GAGTTCTCCATCACCGGCTCCACTTACGCTCCAGAGGGAGAGGTCTTGAAGAATGATAAG CCAGTCCGGCCAGGGCAGTATGACGGGCTGGTGGAGCTGGCCACCATCTGTGCCCTCTGC AATGACTCCTCCTTGGACTTCAACGAGGCCAAAGGTGTCTATGAGAAGGTCGGCGAGGCC ACCGAGACAGCACTCACCACCCTGGTGGAGAAGATGAATGTGTTCAACACGGATGTGAGA AGCCTCTCGAAGGTGGAGAGAGCCAACGCCTGCAACTCGGTGATCCGCCAGCTAATGAAG AAGGAATTCACCCTGGAGTTCTCCCGAGACAGAAAGTCCATGTCTGTCTATTGCTCCCCA GCCAAATCTTCCCGGGCTGCTGTGGGCAACAAGATGTTTGTCAAGGGTGCCCCTGAGGGC GTCATCGACCGCTGTAACTATGTGCGAGTTGGCACCACCCGGGTGCCACTGACGGGGCCG GTGAAGGAAAAGATCATGGCGGTGATCAAGGAGTGGGGCACTGGCCGGGACACCCTGCGC TGCTTGGCCCTGGCCACCCGGGACACCCCCCCGAAGCGAGAGGAAATGGTCCTGGATGAC TCTGCCAGGTTCCTGGAGTATGAGACGGACCTGACATTCGTGGGTGTAGTGGGCATGCTG GACCCTCCGCGCAAGGAGGTCACGGGCTCCATCCAGCTGTGCCGTGACGCCGGGATCCGG GTGATCATGATCACTGGGGACAACAAGGGCACAGCCATTGCCATCTGCCGGCGAATTGGC ATCTTTGGGGAGAACGAGGAGGTGGCCGATCGCGCCTACACGGGCCGAGAGTTCGACGAC CTGCCCCTGGCTGAACAGCGGGAAGCCTGCCGACGTGCCTGCTGCTTCGCCCGTGTGGAG CCCTCGCACAAGTCCAAGATTGTGGAGTACCTGCAGTCCTACGATGAGATCACAGCCATG ACAGGTGATGGCGTCAATGACGCCCCTGCCCTGAAGAAGGCTGAGATTGGCATTGCCATG GGATCTGGCACTGCCGTGGCCAAGACTGCCTCTGAGATGGTGCTGGCTGACGACAACTTC TCCACCATCGTAGCTGCTGTGGAGGAGGGCCGCGCCATCTACAACAACATGAAGCAGTTC ATCCGCTACCTCATTTCCTCCAACGTGGGCGAGGTGGTCTGTATCTTCCTGACCGCTGCC CTGGGGCTGCCTGAGGCCCTGATCCCGGTGCAGCTGCTATGGGTGAACTTGGTGACCGAC GGGCTCCCAGCCACAGCCCTGGGCTTCAACCCACCAGACCTGGACATCATGGACCGCCCC CCCCGGAGCCCCAAGGAGCCCCTCATCAGTGGCTGGCTCTTCTTCCGCTACATGGCAATC GGGGGCTATGTGGGTGCAGCCACCGTGGGAGCAGCTGCCTGGTGGTTCCTGTACGCTGAG GATGGGCCTCATGTCAACTACAGCCAGCTGACTCACTTCATGCAGTGCACCGAGGACAAC ACCCACTTTGAGGGCATAGACTGTGAGGTCTTCGAGGCCCCCGAGCCCATGACCATGGCC CTGTCCGTGCTGGTGACCATCGAGATGTGCAATGCACTGAACAGCCTGTCCGAGAACCAG TCCCTGCTGCGGATGCCACCCTGGGTGAACATCTGGCTGCTGGGCTCCATCTGCCTCTCC ATGTCCCTGCACTTCCTCATCCTCTATGTTGACCCCCTGCCGATGATCTTCAAGCTCCGG GCCCTGGACCTCACCCAGTGGCTCATGGTCCTCAAGATCTCACTGCCAGTCATTGGGCTC GACGAAATCCTCAAGTTCGTTGCTCGGAACTACCTAGAGGATCCAGAAGATGAAAGAAGG AAGTGA |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Number of Residues | 1001 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Weight | 109282.355 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Theoretical pI | 5.175 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pfam Domain Function | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Signals | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transmembrane Regions | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Sequence |
>Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 MEAAHAKTTEECLAYFGVSETTGLTPDQVKRNLEKYGLNELPAEEGKTLWELVIEQFEDL LVRILLLAACISFVLAWFEEGEETITAFVEPFVILLILIANAIVGVWQERNAENAIEALK EYEPEMGKVYRADRKSVQRIKARDIVPGDIVEVAVGDKVPADIRILAIKSTTLRVDQSIL TGESVSVIKHTEPVPDPRAVNQDKKNMLFSGTNIAAGKALGIVATTGVGTEIGKIRDQMA ATEQDKTPLQQKLDEFGEQLSKVISLICVAVWLINIGHFNDPVHGGSWFRGAIYYFKIAV ALAVAAIPEGLPAVITTCLALGTRRMAKKNAIVRSLPSVETLGCTSVICSDKTGTLTTNQ MSVCKMFIIDKVDGDICLLNEFSITGSTYAPEGEVLKNDKPVRPGQYDGLVELATICALC NDSSLDFNEAKGVYEKVGEATETALTTLVEKMNVFNTDVRSLSKVERANACNSVIRQLMK KEFTLEFSRDRKSMSVYCSPAKSSRAAVGNKMFVKGAPEGVIDRCNYVRVGTTRVPLTGP VKEKIMAVIKEWGTGRDTLRCLALATRDTPPKREEMVLDDSARFLEYETDLTFVGVVGML DPPRKEVTGSIQLCRDAGIRVIMITGDNKGTAIAICRRIGIFGENEEVADRAYTGREFDD LPLAEQREACRRACCFARVEPSHKSKIVEYLQSYDEITAMTGDGVNDAPALKKAEIGIAM GSGTAVAKTASEMVLADDNFSTIVAAVEEGRAIYNNMKQFIRYLISSNVGEVVCIFLTAA LGLPEALIPVQLLWVNLVTDGLPATALGFNPPDLDIMDRPPRSPKEPLISGWLFFRYMAI GGYVGAATVGAAAWWFLYAEDGPHVNYSQLTHFMQCTEDNTHFEGIDCEVFEAPEPMTMA LSVLVTIEMCNALNSLSENQSLLRMPPWVNIWLLGSICLSMSLHFLILYVDPLPMIFKLR ALDLTQWLMVLKISLPVIGLDEILKFVARNYLEDPEDERRK |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GenBank ID Protein | 27886529 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProtKB/Swiss-Prot ID | O14983 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProtKB/Swiss-Prot Entry Name | AT2A1_HUMAN | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB IDs | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GenBank Gene ID | NM_173201.3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GeneCard ID | ATP2A1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GenAtlas ID | ATP2A1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HGNC ID | HGNC:811 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||