| Identification |
|---|
| HMDB Protein ID
| CDBP01279 |
| Secondary Accession Numbers
| Not Available |
| Name
| Protein kinase C beta type |
| Description
| Not Available |
| Synonyms
|
- PKC-B
- PKC-beta
|
| Gene Name
| PRKCB |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in protein serine/threonine kinase activity |
| Specific Function
| Calcium-activated and phospholipid-dependent serine/threonine-protein kinase involved in various processes such as regulation of the B-cell receptor (BCR) signalosome, apoptosis and transcription regulation. Plays a key role in B-cell activation and function by regulating BCR-induced NF-kappa-B activation and B-cell suvival. Required for recruitment and activation of the IKK kinase to lipid rafts and mediates phosphorylation of CARD11/CARMA1 at 'Ser-559', 'Ser-644' and 'Ser- 652', leading to activate the NF-kappa-B signaling. Involved in apoptosis following oxidative damage:in case of oxidative conditions, specifically phosphorylates 'Ser-36' of isoform p66Shc of SHC1, leading to mitochondrial accumulation of p66Shc, where p66Shc acts as a reactive oxygen species producer. Acts as a coactivator of androgen receptor (ANDR)-dependent transcription, by being recruited to ANDR target genes and specifically mediating phosphorylation of 'Thr-6' of histone H3 (H3T6ph), a specific tag for epigenetic transcriptional activation that prevents demethylation of histone H3 'Lys-4' (H3K4me) by LSD1/KDM1A. Also involved in triglyceride homeostasis. Serves as the receptor for phorbol esters, a class of tumor promoters |
| GO Classification
|
| Function |
| kinase activity |
| nucleoside binding |
| purine nucleoside binding |
| adenyl nucleotide binding |
| adenyl ribonucleotide binding |
| atp binding |
| protein kinase c activity |
| protein kinase activity |
| ion binding |
| protein serine/threonine kinase activity |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| transition metal ion binding |
| zinc ion binding |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| Process |
| signaling pathway |
| phosphorylation |
| phosphorus metabolic process |
| phosphate metabolic process |
| metabolic process |
| cellular metabolic process |
| protein amino acid phosphorylation |
| intracellular signaling pathway |
| signaling |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
- Peripheral membrane protein
- Membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Chlorphenamine H1-Antihistamine Action | 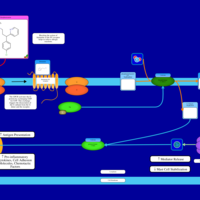   | Not Available | | Pheniramine H1-Antihistamine Action | 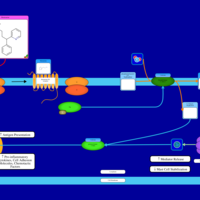 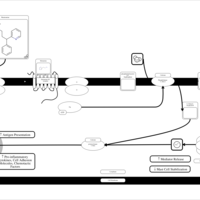 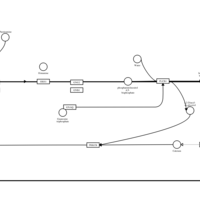 | Not Available | | Dexchlorpheniramine H1-Antihistamine Action | 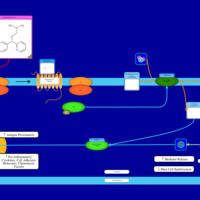 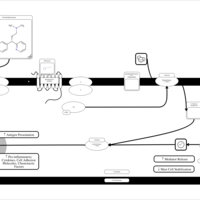 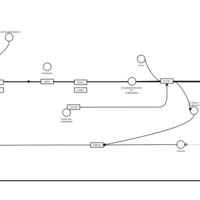 | Not Available | | Brompheniramine H1-Antihistamine Action | 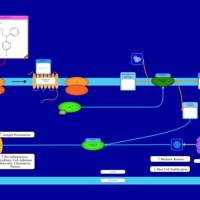 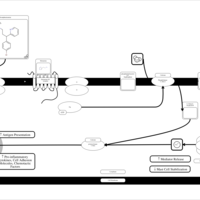 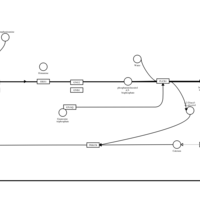 | Not Available | | Dexbrompheniramine H1-Antihistamine Action | 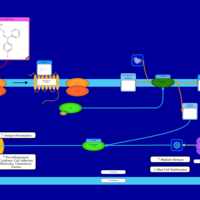 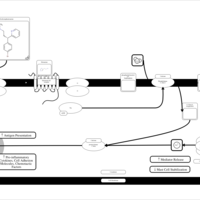 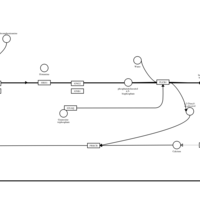 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| Chromosome:1 |
| Locus
| 16p11.2 |
| SNPs
| PRKCB |
| Gene Sequence
|
>2016 bp
ATGGCTGACCCGGCTGCGGGGCCGCCGCCGAGCGAGGGCGAGGAGAGCACCGTGCGCTTC
GCCCGCAAAGGCGCCCTCCGGCAGAAGAACGTGCATGAGGTCAAGAACCACAAATTCACC
GCCCGCTTCTTCAAGCAGCCCACCTTCTGCAGCCACTGCACCGACTTCATCTGGGGCTTC
GGGAAGCAGGGATTCCAGTGCCAAGTTTGCTGCTTTGTGGTGCACAAGCGGTGCCATGAA
TTTGTCACATTCTCCTGCCCTGGCGCTGACAAGGGTCCAGCCTCCGATGACCCCCGCAGC
AAACACAAGTTTAAGATCCACACGTACTCCAGCCCCACGTTTTGTGACCACTGTGGGTCA
CTGCTGTATGGACTCATCCACCAGGGGATGAAATGTGACACCTGCATGATGAATGTGCAC
AAGCGCTGCGTGATGAATGTTCCCAGCCTGTGTGGCACGGACCACACGGAGCGCCGCGGC
CGCATCTACATCCAGGCCCACATCGACAGGGACGTCCTCATTGTCCTCGTAAGAGATGCT
AAAAACCTTGTACCTATGGACCCCAATGGCCTGTCAGATCCCTACGTAAAACTGAAACTG
ATTCCCGATCCCAAAAGTGAGAGCAAACAGAAGACCAAAACCATCAAATGCTCCCTCAAC
CCTGAGTGGAATGAGACATTTAGATTTCAGCTGAAAGAATCGGACAAAGACAGAAGACTG
TCAGTAGAGATTTGGGATTGGGATTTGACCAGCAGGAATGACTTCATGGGATCTTTGTCC
TTTGGGATTTCTGAACTTCAGAAAGCCAGTGTTGATGGCTGGTTTAAGTTACTGAGCCAG
GAGGAAGGCGAGTACTTCAATGTGCCTGTGCCACCAGAAGGAAGTGAGGCCAATGAAGAA
CTGCGGCAGAAATTTGAGAGGGCCAAGATCAGTCAGGGAACCAAGGTCCCGGAAGAAAAG
ACGACCAACACTGTCTCCAAATTTGACAACAATGGCAACAGAGACCGGATGAAACTGACC
GATTTTAACTTCCTAATGGTGCTGGGGAAAGGCAGCTTTGGCAAGGTCATGCTTTCAGAA
CGAAAAGGCACAGATGAGCTCTATGCTGTGAAGATCCTGAAGAAGGACGTTGTGATCCAA
GATGATGACGTGGAGTGCACTATGGTGGAGAAGCGGGTGTTGGCCCTGCCTGGGAAGCCG
CCCTTCCTGACCCAGCTCCACTCCTGCTTCCAGACCATGGACCGCCTGTACTTTGTGATG
GAGTACGTGAATGGGGGCGACCTCATGTATCACATCCAGCAAGTCGGCCGGTTCAAGGAG
CCCCATGCTGTATTTTACGCTGCAGAAATTGCCATCGGTCTGTTCTTCTTACAGAGTAAG
GGCATCATTTACCGTGACCTAAAACTTGACAACGTGATGCTCGATTCTGAGGGACACATC
AAGATTGCCGATTTTGGCATGTGTAAGGAAAACATCTGGGATGGGGTGACAACCAAGACA
TTCTGTGGCACTCCAGACTACATCGCCCCCGAGATAATTGCTTATCAGCCCTATGGGAAG
TCCGTGGATTGGTGGGCATTTGGAGTCCTGCTGTATGAAATGTTGGCTGGGCAGGCACCC
TTTGAAGGGGAGGATGAAGATGAACTCTTCCAATCCATCATGGAACACAACGTAGCCTAT
CCCAAGTCTATGTCCAAGGAAGCTGTGGCCATCTGCAAAGGGCTGATGACCAAACACCCA
GGCAAACGTCTGGGTTGTGGACCTGAAGGCGAACGTGATATCAAAGAGCATGCATTTTTC
CGGTATATTGATTGGGAGAAACTTGAACGCAAAGAGATCCAGCCCCCTTATAAGCCAAAA
GCTAGAGACAAGAGAGACACCTCCAACTTCGACAAAGAGTTCACCAGACAGCCTGTGGAA
CTGACCCCCACTGATAAACTCTTCATCATGAACTTGGACCAAAATGAATTTGCTGGCTTC
TCTTATACTAACCCAGAGTTTGTCATTAATGTGTAG
|
| Protein Properties |
|---|
| Number of Residues
| 671 |
| Molecular Weight
| 76868.4 |
| Theoretical pI
| 7.0 |
| Pfam Domain Function
|
|
| Signals
|
|
|
Transmembrane Regions
|
|
| Protein Sequence
|
>Protein kinase C beta type
MADPAAGPPPSEGEESTVRFARKGALRQKNVHEVKNHKFTARFFKQPTFCSHCTDFIWGF
GKQGFQCQVCCFVVHKRCHEFVTFSCPGADKGPASDDPRSKHKFKIHTYSSPTFCDHCGS
LLYGLIHQGMKCDTCMMNVHKRCVMNVPSLCGTDHTERRGRIYIQAHIDRDVLIVLVRDA
KNLVPMDPNGLSDPYVKLKLIPDPKSESKQKTKTIKCSLNPEWNETFRFQLKESDKDRRL
SVEIWDWDLTSRNDFMGSLSFGISELQKASVDGWFKLLSQEEGEYFNVPVPPEGSEANEE
LRQKFERAKISQGTKVPEEKTTNTVSKFDNNGNRDRMKLTDFNFLMVLGKGSFGKVMLSE
RKGTDELYAVKILKKDVVIQDDDVECTMVEKRVLALPGKPPFLTQLHSCFQTMDRLYFVM
EYVNGGDLMYHIQQVGRFKEPHAVFYAAEIAIGLFFLQSKGIIYRDLKLDNVMLDSEGHI
KIADFGMCKENIWDGVTTKTFCGTPDYIAPEIIAYQPYGKSVDWWAFGVLLYEMLAGQAP
FEGEDEDELFQSIMEHNVAYPKSMSKEAVAICKGLMTKHPGKRLGCGPEGERDIKEHAFF
RYIDWEKLERKEIQPPYKPKARDKRDTSNFDKEFTRQPVELTPTDKLFIMNLDQNEFAGF
SYTNPEFVINV
|
| External Links |
|---|
| GenBank ID Protein
| 47157322 |
| UniProtKB/Swiss-Prot ID
| P05771 |
| UniProtKB/Swiss-Prot Entry Name
| KPCB_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| NM_212535.2 |
| GeneCard ID
| PRKCB |
| GenAtlas ID
| PRKCB |
| HGNC ID
| HGNC:9395 |
| References |
|---|
| General References
| Not Available |