| Biological Process |
| cellular iron ion homeostasis |
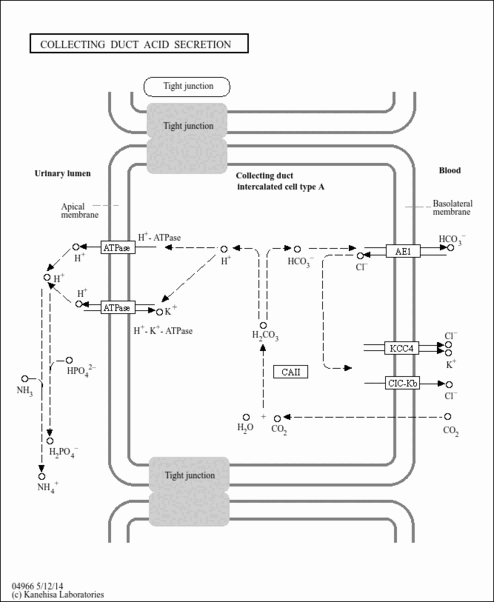
| ATP hydrolysis coupled proton transport |
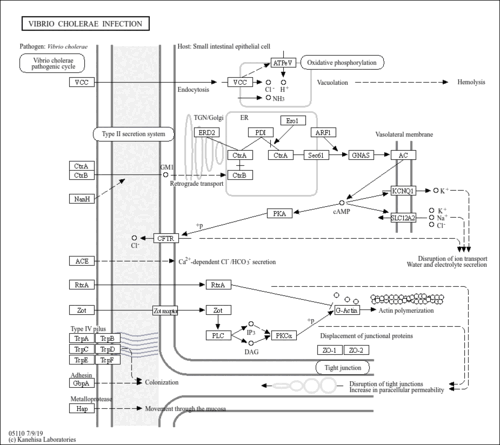
| interaction with host |
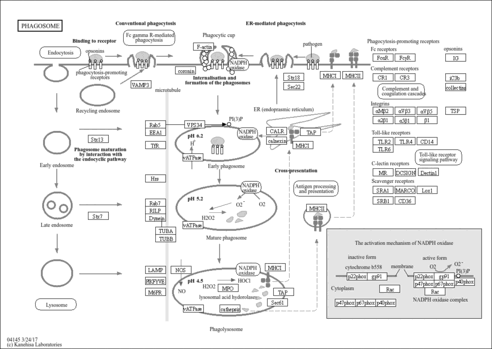
| phagosome maturation |
| transferrin transport |
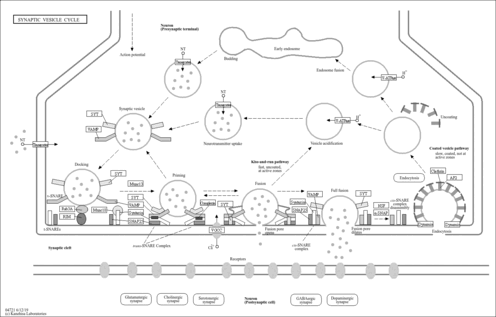
| transmembrane transport |
| insulin receptor signaling pathway |
| Cellular Component |
| cytosol |
| mitochondrion |
| proton-transporting two-sector ATPase complex |
| proton-transporting V-type ATPase, V1 domain |
| microvillus |
| apical plasma membrane |
| integral to plasma membrane |
| Component |
| macromolecular complex |
| protein complex |
| proton-transporting two-sector atpase complex |
| proton-transporting two-sector atpase complex, catalytic domain |
| proton-transporting v-type atpase, v1 domain |
| Function |
| inorganic cation transmembrane transporter activity |
| monovalent inorganic cation transmembrane transporter activity |
| binding |
| hydrogen ion transmembrane transporter activity |
| catalytic activity |
| transporter activity |
| hydrolase activity |
| hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances |
| nucleoside binding |
| hydrolase activity, acting on acid anhydrides |
| purine nucleoside binding |
| adenyl nucleotide binding |
| adenyl ribonucleotide binding |
| atp binding |
| proton-transporting atpase activity, rotational mechanism |
| hydrogen ion transporting atp synthase activity, rotational mechanism |
| transmembrane transporter activity |
| substrate-specific transmembrane transporter activity |
| ion transmembrane transporter activity |
| cation transmembrane transporter activity |
| Molecular Function |
| ATP binding |
| hydrogen ion transporting ATP synthase activity, rotational mechanism |
| proton-transporting ATPase activity, rotational mechanism |
| Process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside phosphate metabolic process |
| nucleotide metabolic process |
| atp synthesis coupled proton transport |
| purine nucleoside triphosphate metabolic process |
| purine ribonucleoside triphosphate metabolic process |
| atp metabolic process |
| hydrogen transport |
| purine nucleotide metabolic process |
| proton transport |
| purine nucleotide biosynthetic process |
| establishment of localization |
| purine nucleoside triphosphate biosynthetic process |
| purine ribonucleoside triphosphate biosynthetic process |
| transport |
| metabolic process |
| atp biosynthetic process |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |