| Identification |
|---|
| HMDB Protein ID
| CDBP01210 |
| Secondary Accession Numbers
| Not Available |
| Name
| Polyribonucleotide nucleotidyltransferase 1, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- 3'-5' RNA exonuclease OLD35
- PNPase 1
- PNPase old-35
- Polynucleotide phosphorylase 1
- Polynucleotide phosphorylase-like protein
|
| Gene Name
| PNPT1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in 3'-5'-exoribonuclease activity |
| Specific Function
| RNA-binding protein implicated in numerous RNA metabolic processes. Hydrolyzes single-stranded polyribonucleotides processively in the 3'-to-5' direction. Mitochondrial intermembrane factor with RNA-processing exoribonulease activity. Component of the mitochondrial degradosome (mtEXO) complex, that degrades 3' overhang double-stranded RNA with a 3'-to-5' directionality in an ATP-dependent manner. Required for correct processing and polyadenylation of mitochondrial mRNAs. Plays a role as a cytoplasmic RNA import factor that mediates the translocation of small RNA components, like the 5S RNA, the RNA subunit of ribonuclease P and the mitochondrial RNA-processing (MRP) RNA, into the mitochondrial matrix. Plays a role in mitochondrial morphogenesis and respiration; regulates the expression of the electron transport chain (ETC) components at the mRNA and protein levels. In the cytoplasm, shows a 3'-to-5' exoribonuclease mediating mRNA degradation activity; degrades c-myc mRNA upon treatment with IFNB1/IFN-beta, resulting in a growth arrest in melanoma cells. Regulates the stability of specific mature miRNAs in melanoma cells; specifically and selectively degrades miR-221, preferentially. Plays also a role in RNA cell surveillance by cleaning up oxidized RNAs. Binds to the RNA subunit of ribonuclease P, MRP RNA and miR-221 microRNA.
|
| GO Classification
|
| Biological Process |
| nuclear polyadenylation-dependent mRNA catabolic process |
| positive regulation of miRNA catabolic process |
| positive regulation of mitochondrial RNA catabolic process |
| positive regulation of mRNA catabolic process |
| regulation of cellular senescence |
| rRNA import into mitochondrion |
| regulation of cellular respiration |
| protein homotrimerization |
| cellular response to oxidative stress |
| mitochondrion morphogenesis |
| negative regulation of growth |
| cellular response to interferon-beta |
| mitochondrial mRNA catabolic process |
| mitochondrial mRNA polyadenylation |
| mitochondrial RNA 3'-end processing |
| mitochondrial RNA 5'-end processing |
| mitotic cell cycle arrest |
| Cellular Component |
| plasma membrane |
| mitochondrial degradosome |
| mitochondrial intermembrane space |
| Component |
| mitochondrion |
| organelle |
| membrane-bounded organelle |
| intracellular membrane-bounded organelle |
| Function |
| hydrolase activity, acting on ester bonds |
| binding |
| catalytic activity |
| hydrolase activity |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| nucleotidyltransferase activity |
| polyribonucleotide nucleotidyltransferase activity |
| exonuclease activity |
| 3'-5' exonuclease activity |
| 3'-5'-exoribonuclease activity |
| nucleic acid binding |
| nuclease activity |
| rna binding |
| Molecular Function |
| poly(G) RNA binding |
| poly(U) RNA binding |
| polyribonucleotide nucleotidyltransferase activity |
| 3'-5'-exoribonuclease activity |
| miRNA binding |
| Process |
| rna metabolic process |
| metabolic process |
| macromolecule metabolic process |
| cellular macromolecule metabolic process |
| rna processing |
| rna catabolic process |
| mrna catabolic process |
|
| Cellular Location
|
- Mitochondrion
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Pyrimidine Metabolism |    |  | | RNA degradation | Not Available | 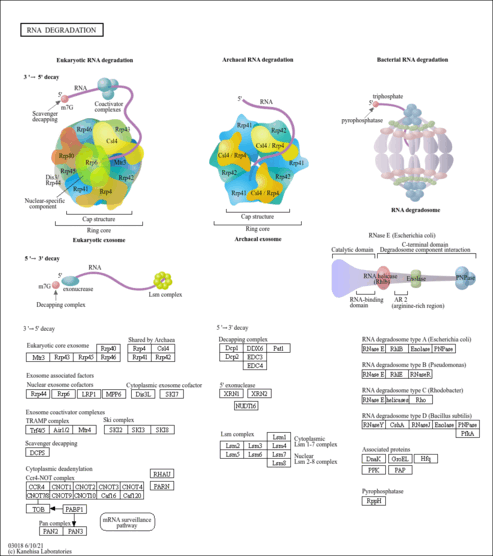 |
|
| Gene Properties |
|---|
| Chromosome Location
| 2 |
| Locus
| 2p15 |
| SNPs
| PNPT1 |
| Gene Sequence
|
>2352 bp
ATGGCGGCCTGCAGGTACTGCTGCTCGTGCCTCCGGCTCCGGCCCCTGAGCGATGGTCCT
TTCCTTCTGCCACGGCGGGATCGGGCACTCACCCAGTTGCAAGTGCGAGCACTATGGAGT
AGCGCAGGGTCTCGAGCTGTGGCCGTGGACTTAGGCAACAGGAAATTAGAAATATCTTCT
GGAAAGCTGGCCAGATTTGCAGATGGCTCTGCTGTAGTACAGTCAGGTGACACTGCAGTA
ATGGTCACAGCGGTCAGTAAAACAAAACCTTCCCCTTCCCAGTTTATGCCTTTGGTGGTT
GACTACAGACAAAAAGCTGCTGCAGCAGGTAGAATTCCCACAAACTATCTGAGAAGAGAG
ATTGGTACTTCTGATAAAGAAATTCTAACAAGTCGAATAATAGATCGTTCAATTAGACCG
CTCTTTCCAGCTGGCTACTTCTATGATACACAGGTTCTGTGTAATCTGTTAGCAGTAGAT
GGTGTAAATGAGCCTGATGTCCTAGCAATTAATGGCGCTTCCGTAGCCCTCTCATTATCA
GATATTCCTTGGAATGGACCTGTTGGGGCAGTACGAATAGGAATAATTGATGGAGAATAT
GTTGTTAACCCAACAAGAAAAGAAATGTCTTCTAGTACTTTAAATTTAGTGGTTGCTGGA
GCACCTAAAAGTCAGATTGTCATGTTGGAAGCCTCTGCAGAGAACATTTTACAGCAGGAC
TTTTGCCATGCTATCAAAGTGGGAGTGAAATATACCCAACAAATAATTCAGGGCATTCAG
CAGTTGGTAAAAGAAACTGGTGTTACCAAGAGGACACCTCAGAAGTTATTTACCCCTTCG
CCAGAGATTGTGAAATATACTCATAAACTTGCTATGGAGAGACTCTATGCAGTTTTTACA
GATTACGAGCATGACAAAGTTTCCAGAGATGAAGCTGTTAACAAAATAAGATTAGATACG
GAGGAACAACTAAAAGAAAAATTTCCAGAAGCCGATCCATATGAAATAATAGAATCCTTC
AATGTTGTTGCAAAGGAAGTTTTTAGAAGTATTGTTTTGAATGAATACAAAAGGTGCGAT
GGTCGGGATTTGACTTCACTTAGGAATGTAAGTTGTGAGGTAGATATGTTTAAAACCCTT
CATGGATCAGCATTATTTCAAAGAGGACAAACACAGGTGCTTTGTACCGTTACATTTGAT
TCATTAGAATCTGGTATTAAGTCAGATCAAGTTATAACAGCTATAAATGGGATAAAAGAT
AAAAATTTCATGCTGCACTACGAGTTTCCTCCTTATGCAACTAATGAAATTGGCAAAGTC
ACTGGTTTAAATAGAAGAGAACTTGGGCATGGTGCTCTTGCTGAGAAAGCTTTGTATCCT
GTTATTCCCCGAGATTTTCCTTTCACCATAAGAGTTACATCTGAAGTCCTAGAGTCAAAT
GGGTCATCTTCTATGGCATCTGCATGTGGCGGAAGTTTAGCATTAATGGATTCAGGGGTT
CCAATTTCATCTGCTGTTGCAGGCGTAGCAATAGGATTGGTCACCAAAACCGATCCTGAG
AAGGGTGAAATAGAAGATTATCGTTTGCTGACAGATATTTTGGGAATTGAAGATTACAAT
GGTGACATGGACTTCAAAATAGCTGGCACTAATAAAGGAATAACTGCATTACAGGCTGAT
ATTAAATTACCTGGAATACCAATAAAAATTGTGATGGAGGCTATTCAACAAGCTTCAGTG
GCAAAAAAGGAGATATTACAGATCATGAACAAAACTATTTCAAAACCTCGAGCATCTAGA
AAAGAAAATGGACCTGTTGTAGAAACTGTTCAGGTTCCATTATCAAAACGAGCAAAATTT
GTTGGACCTGGTGGCTATAACTTAAAAAAACTTCAGGCTGAAACAGGTGTAACTATTAGT
CAGGTGGATGAAGAAACGTTTTCTGTATTTGCACCAACACCCAGTGCTATGCATGAGGCA
AGAGACTTCATTACTGAAATCTGCAAGGATGATCAGGAGCAGCAATTAGAATTTGGAGCA
GTATATACCGCCACAATAACTGAAATCAGAGATACTGGTGTAATGGTAAAATTATATCCA
AATATGACTGCGGTACTGCTTCATAACACACAACTTGATCAACGAAAGATTAAACATCCT
ACTGCCCTAGGATTAGAAGTTGGCCAAGAAATTCAGGTGAAATACTTTGGACGTGACCCA
GCCGATGGAAGAATGAGGCTTTCTCGAAAAGTGCTTCAGTCGCCAGCTACAACCGTGGTC
AGAACTTTGAATGACAGAAGTAGTATTGTAATGGGAGAACCTATTTCACAGTCATCATCT
AATTCTCAGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 783 |
| Molecular Weight
| 85949.84 |
| Theoretical pI
| 7.769 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Polyribonucleotide nucleotidyltransferase 1, mitochondrial
MAACRYCCSCLRLRPLSDGPFLLPRRDRALTQLQVRALWSSAGSRAVAVDLGNRKLEISS
GKLARFADGSAVVQSGDTAVMVTAVSKTKPSPSQFMPLVVDYRQKAAAAGRIPTNYLRRE
IGTSDKEILTSRIIDRSIRPLFPAGYFYDTQVLCNLLAVDGVNEPDVLAINGASVALSLS
DIPWNGPVGAVRIGIIDGEYVVNPTRKEMSSSTLNLVVAGAPKSQIVMLEASAENILQQD
FCHAIKVGVKYTQQIIQGIQQLVKETGVTKRTPQKLFTPSPEIVKYTHKLAMERLYAVFT
DYEHDKVSRDEAVNKIRLDTEEQLKEKFPEADPYEIIESFNVVAKEVFRSIVLNEYKRCD
GRDLTSLRNVSCEVDMFKTLHGSALFQRGQTQVLCTVTFDSLESGIKSDQVITAINGIKD
KNFMLHYEFPPYATNEIGKVTGLNRRELGHGALAEKALYPVIPRDFPFTIRVTSEVLESN
GSSSMASACGGSLALMDSGVPISSAVAGVAIGLVTKTDPEKGEIEDYRLLTDILGIEDYN
GDMDFKIAGTNKGITALQADIKLPGIPIKIVMEAIQQASVAKKEILQIMNKTISKPRASR
KENGPVVETVQVPLSKRAKFVGPGGYNLKKLQAETGVTISQVDEETFSVFAPTPSAMHEA
RDFITEICKDDQEQQLEFGAVYTATITEIRDTGVMVKLYPNMTAVLLHNTQLDQRKIKHP
TALGLEVGQEIQVKYFGRDPADGRMRLSRKVLQSPATTVVRTLNDRSSIVMGEPISQSSS
NSQ
|
| External Links |
|---|
| GenBank ID Protein
| 62988884 |
| UniProtKB/Swiss-Prot ID
| Q8TCS8 |
| UniProtKB/Swiss-Prot Entry Name
| PNPT1_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AC015982 |
| GeneCard ID
| PNPT1 |
| GenAtlas ID
| PNPT1 |
| HGNC ID
| HGNC:23166 |
| References |
|---|
| General References
| Not Available |

