| Identification |
|---|
| HMDB Protein ID
| CDBP01133 |
| Secondary Accession Numbers
| Not Available |
| Name
| ATP synthase subunit epsilon, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- ATPase subunit epsilon
|
| Gene Name
| ATP5E |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in hydrogen ion transporting ATP synthase activity, rotational mechanism |
| Specific Function
| Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core, and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Part of the complex F(1) domain and of the central stalk which is part of the complex rotary element. Rotation of the central stalk against the surrounding alpha(3)beta(3) subunits leads to hydrolysis of ATP in three separate catalytic sites on the beta subunits |
| GO Classification
|
| Component |
| membrane part |
| mitochondrial proton-transporting atp synthase complex, catalytic core f(1) |
| mitochondrial membrane part |
| cell part |
| Function |
| proton-transporting atpase activity, rotational mechanism |
| hydrogen ion transporting atp synthase activity, rotational mechanism |
| transmembrane transporter activity |
| substrate-specific transmembrane transporter activity |
| ion transmembrane transporter activity |
| cation transmembrane transporter activity |
| inorganic cation transmembrane transporter activity |
| monovalent inorganic cation transmembrane transporter activity |
| hydrogen ion transmembrane transporter activity |
| transporter activity |
| Process |
| purine ribonucleoside triphosphate biosynthetic process |
| metabolic process |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside phosphate metabolic process |
| nucleotide metabolic process |
| atp synthesis coupled proton transport |
| atp biosynthetic process |
| purine nucleotide metabolic process |
| purine nucleotide biosynthetic process |
| purine nucleoside triphosphate biosynthetic process |
|
| Cellular Location
|
- Mitochondrion
- Mitochondrion inner membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Mitochondrial Electron Transport Chain | 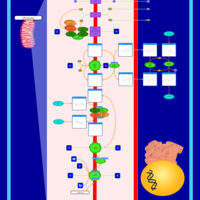 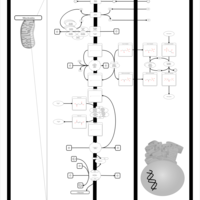 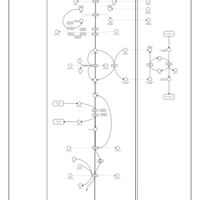 |  |
|
| Gene Properties |
|---|
| Chromosome Location
| Chromosome:2 |
| Locus
| 20q13.32 |
| SNPs
| ATP5E |
| Gene Sequence
|
>156 bp
ATGGTGGCCTACTGGAGACAGGCTGGACTCAGCTACATCCGATACTCCCAGATCTGTGCA
AAAGCAGTGAGAGATGCACTGAAGACAGAATTCAAAGCAAATGCTGAGAAGACTTCTGGC
AGCAACGTAAAAATTGTGAAAGTAAAGAAGGAATAA
|
| Protein Properties |
|---|
| Number of Residues
| 51 |
| Molecular Weight
| 5779.7 |
| Theoretical pI
| 10.53 |
| Pfam Domain Function
|
|
| Signals
|
|
|
Transmembrane Regions
|
|
| Protein Sequence
|
>ATP synthase subunit epsilon, mitochondrial
MVAYWRQAGLSYIRYSQICAKAVRDALKTEFKANAEKTSGSNVKIVKVKKE
|
| External Links |
|---|
| GenBank ID Protein
| 8117712 |
| UniProtKB/Swiss-Prot ID
| P56381 |
| UniProtKB/Swiss-Prot Entry Name
| ATP5E_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF052955 |
| GeneCard ID
| ATP5E |
| GenAtlas ID
| ATP5E |
| HGNC ID
| HGNC:838 |
| References |
|---|
| General References
| Not Available |
