| Identification |
|---|
| HMDB Protein ID
| CDBP01021 |
| Secondary Accession Numbers
| Not Available |
| Name
| Alpha-enolase |
| Description
| Not Available |
| Synonyms
|
- 2-phospho-D-glycerate hydro-lyase
- C-myc promoter-binding protein
- Enolase 1
- MBP-1
- MPB-1
- NNE
- Non-neural enolase
- Phosphopyruvate hydratase
- Plasminogen-binding protein
|
| Gene Name
| ENO1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in magnesium ion binding |
| Specific Function
| Multifunctional enzyme that, as well as its role in glycolysis, plays a part in various processes such as growth control, hypoxia tolerance and allergic responses. May also function in the intravascular and pericellular fibrinolytic system due to its ability to serve as a receptor and activator of plasminogen on the cell surface of several cell-types such as leukocytes and neurons. Stimulates immunoglobulin production.
MBP1 binds to the myc promoter and acts as a transcriptional repressor. May be a tumor suppressor.
|
| GO Classification
|
| Biological Process |
| small molecule metabolic process |
| glycolysis |
| response to virus |
| gluconeogenesis |
| negative regulation of transcription from RNA polymerase II promoter |
| negative regulation of cell growth |
| transcription, DNA-dependent |
| Cellular Component |
| cytosol |
| M band |
| phosphopyruvate hydratase complex |
| extracellular vesicular exosome |
| plasma membrane |
| nucleus |
| Component |
| macromolecular complex |
| protein complex |
| phosphopyruvate hydratase complex |
| Function |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| magnesium ion binding |
| lyase activity |
| carbon-oxygen lyase activity |
| hydro-lyase activity |
| phosphopyruvate hydratase activity |
| Molecular Function |
| magnesium ion binding |
| phosphopyruvate hydratase activity |
| sequence-specific DNA binding transcription factor activity |
| transcription corepressor activity |
| DNA binding |
| Process |
| small molecule metabolic process |
| alcohol metabolic process |
| monosaccharide metabolic process |
| hexose metabolic process |
| glucose metabolic process |
| glucose catabolic process |
| glycolysis |
| metabolic process |
|
| Cellular Location
|
- Isoform MBP-1:Nucleus
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Glycolysis / Gluconeogenesis | Not Available |  | | RNA degradation | Not Available |  | | HIF-1 signaling pathway | Not Available |  | | Glycolysis |   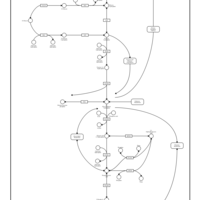 |  | | Glycogenosis, Type VII. Tarui disease | 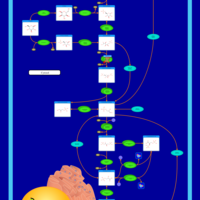 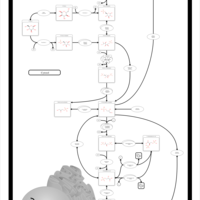 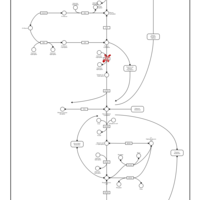 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 1 |
| Locus
| 1p36.2 |
| SNPs
| ENO1 |
| Gene Sequence
|
>1305 bp
ATGTCTATTCTCAAGATCCATGCCAGGGAGATCTTTGACTCTCGCGGGAATCCCACTGTT
GAGGTTGATCTCTTCACCTCAAAAGGTCTCTTCAGAGCTGCTGTGCCCAGTGGTGCTTCA
ACTGGTATCTATGAGGCCCTAGAGCTCCGGGACAATGATAAGACTCGCTATATGGGGAAG
GGTGTCTCAAAGGCTGTTGAGCACATCAATAAAACTATTGCGCCTGCCCTGGTTAGCAAG
AAACTGAACGTCACAGAACAAGAGAAGATTGACAAACTGATGATCGAGATGGATGGAACA
GAAAATAAATCTAAGTTTGGTGCGAACGCCATTCTGGGGGTGTCCCTTGCCGTCTGCAAA
GCTGGTGCCGTTGAGAAGGGGGTCCCCCTGTACCGCCACATCGCTGACTTGGCTGGCAAC
TCTGAAGTCATCCTGCCAGTCCCGGCGTTCAATGTCATCAATGGCGGTTCTCATGCTGGC
AACAAGCTGGCCATGCAGGAGTTCATGATCCTCCCAGTCGGTGCAGCAAACTTCAGGGAA
GCCATGCGCATTGGAGCAGAGGTTTACCACAACCTGAAGAATGTCATCAAGGAGAAATAT
GGGAAAGATGCCACCAATGTGGGGGATGAAGGCGGGTTTGCTCCCAACATCCTGGAGAAT
AAAGAAGGCCTGGAGCTGCTGAAGACTGCTATTGGGAAAGCTGGCTACACTGATAAGGTG
GTCATCGGCATGGACGTAGCGGCCTCCGAGTTCTTCAGGTCTGGGAAGTATGACCTGGAC
TTCAAGTCTCCCGATGACCCCAGCAGGTACATCTCGCCTGACCAGCTGGCTGACCTGTAC
AAGTCCTTCATCAAGGACTACCCAGTGGTGTCTATCGAAGATCCCTTTGACCAGGATGAC
TGGGGAGCTTGGCAGAAGTTCACAGCCAGTGCAGGAATCCAGGTAGTGGGGGATGATCTC
ACAGTGACCAACCCAAAGAGGATCGCCAAGGCCGTGAACGAGAAGTCCTGCAACTGCCTC
CTGCTCAAAGTCAACCAGATTGGCTCCGTGACCGAGTCTCTTCAGGCGTGCAAGCTGGCC
CAGGCCAATGGTTGGGGCGTCATGGTGTCTCATCGTTCGGGGGAGACTGAAGATACCTTC
ATCGCTGACCTGGTTGTGGGGCTGTGCACTGGGCAGATCAAGACTGGTGCCCCTTGCCGA
TCTGAGCGCTTGGCCAAGTACAACCAGCTCCTCAGAATTGAAGAGGAGCTGGGCAGCAAG
GCTAAGTTTGCCGGCAGGAACTTCAGAAACCCCTTGGCCAAGTAA
|
| Protein Properties |
|---|
| Number of Residues
| 434 |
| Molecular Weight
| 36927.84 |
| Theoretical pI
| 6.278 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Alpha-enolase
MSILKIHAREIFDSRGNPTVEVDLFTSKGLFRAAVPSGASTGIYEALELRDNDKTRYMGK
GVSKAVEHINKTIAPALVSKKLNVTEQEKIDKLMIEMDGTENKSKFGANAILGVSLAVCK
AGAVEKGVPLYRHIADLAGNSEVILPVPAFNVINGGSHAGNKLAMQEFMILPVGAANFRE
AMRIGAEVYHNLKNVIKEKYGKDATNVGDEGGFAPNILENKEGLELLKTAIGKAGYTDKV
VIGMDVAASEFFRSGKYDLDFKSPDDPSRYISPDQLADLYKSFIKDYPVVSIEDPFDQDD
WGAWQKFTASAGIQVVGDDLTVTNPKRIAKAVNEKSCNCLLLKVNQIGSVTESLQACKLA
QANGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGSK
AKFAGRNFRNPLAK
|
| External Links |
|---|
| GenBank ID Protein
| 4503571 |
| UniProtKB/Swiss-Prot ID
| P06733 |
| UniProtKB/Swiss-Prot Entry Name
| ENOA_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_001428.2 |
| GeneCard ID
| ENO1 |
| GenAtlas ID
| ENO1 |
| HGNC ID
| HGNC:3350 |
| References |
|---|
| General References
| Not Available |


