| Identification |
|---|
| HMDB Protein ID
| CDBP01002 |
| Secondary Accession Numbers
| Not Available |
| Name
| Retinal guanylyl cyclase 1 |
| Description
| Not Available |
| Synonyms
|
- Guanylate cyclase 2D, retinal
- RETGC-1
- ROS-GC
- Rod outer segment membrane guanylate cyclase
|
| Gene Name
| GUCY2D |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in phosphorus-oxygen lyase activity |
| Specific Function
| Probably plays a specific functional role in the rods and/or cones of photoreceptors. It may be the enzyme involved in the resynthesis of cGMP required for recovery of the dark state after phototransduction.
|
| GO Classification
|
| Biological Process |
| receptor guanylyl cyclase signaling pathway |
| visual perception |
| intracellular signal transduction |
| Cellular Component |
| nuclear outer membrane |
| integral to plasma membrane |
| Function |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| kinase activity |
| nucleoside binding |
| purine nucleoside binding |
| adenyl nucleotide binding |
| adenyl ribonucleotide binding |
| atp binding |
| lyase activity |
| cyclase activity |
| phosphorus-oxygen lyase activity |
| protein kinase activity |
| guanylate cyclase activity |
| binding |
| catalytic activity |
| Molecular Function |
| GTP binding |
| receptor activity |
| ATP binding |
| guanylate cyclase activity |
| protein kinase activity |
| Process |
| cyclic nucleotide biosynthetic process |
| protein amino acid phosphorylation |
| cgmp biosynthetic process |
| phosphorus metabolic process |
| intracellular signaling pathway |
| phosphate metabolic process |
| signaling |
| purine nucleotide metabolic process |
| signaling pathway |
| purine nucleotide biosynthetic process |
| phosphorylation |
| metabolic process |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside phosphate metabolic process |
| nucleotide metabolic process |
| nucleoside monophosphate metabolic process |
| nucleoside monophosphate biosynthetic process |
| cellular metabolic process |
|
| Cellular Location
|
- Membrane
- Single-pass type I membrane protein
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Olfactory transduction | Not Available | 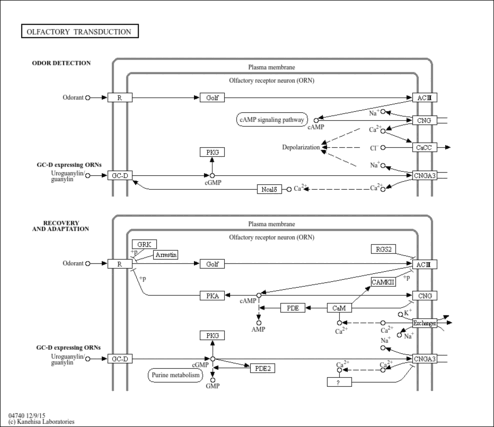 | | Phototransduction | Not Available | 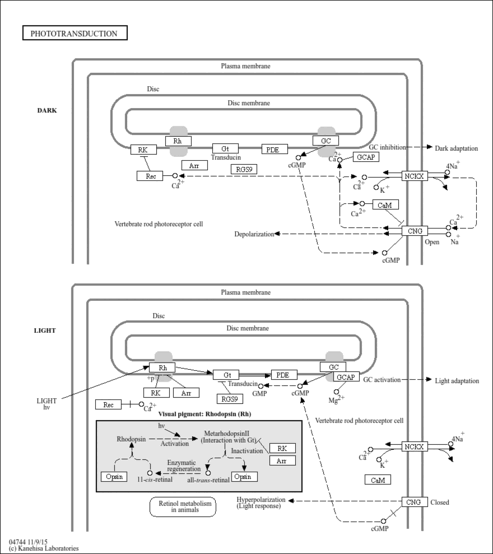 |
|
| Gene Properties |
|---|
| Chromosome Location
| 17 |
| Locus
| 17p13.1 |
| SNPs
| GUCY2D |
| Gene Sequence
|
>3312 bp
ATGACCGCCTGCGCCCGCCGAGCGGGTGGGCTTCCGGACCCCGGGCTCTGCGGTCCCGCG
TGGTGGGCTCCGTCCCTGCCCCGCCTCCCCCGGGCCCTGCCCCGGCTCCCGCTCCTGCTG
CTCCTGCTTCTGCTGCAGCCCCCCGCCCTCTCCGCCGTGTTCACGGTGGGGGTCCTGGGC
CCCTGGGCTTGCGACCCCATCTTCTCTCGGGCTCGCCCGGACCTGGCCGCCCGCCTGGCC
GCCGCCCGCCTGAACCGCGACCCCGGCCTGGCAGGCGGTCCCCGCTTCGAGGTAGCGCTG
CTGCCCGAGCCTTGCCGGACGCCGGGCTCGCTGGGGGCCGTGTCCTCCGCGCTGGCCCGC
GTGTCGGGCCTCGTGGGTCCGGTGAACCCTGCGGCCTGCCGGCCAGCCGAGCTGCTCGCC
GAAGAAGCCGGGATCGCGCTGGTGCCCTGGGGCTGCCCCTGGACGCAGGCGGAGGGCACC
ACGGCCCCTGCCGTGACCCCCGCCGCGGATGCCCTCTACGCCCTGCTTCGCGCATTCGGC
TGGGCGCGCGTGGCCCTGGTCACCGCCCCCCAGGACCTGTGGGTGGAGGCGGGACGCTCA
CTGTCCACGGCACTCAGGGCCCGGGGCCTGCCTGTCGCCTCCGTGACTTCCATGGAGCCC
TTGGACCTGTCTGGAGCCCGGGAGGCCCTGAGGAAGGTTCGGGACGGGCCCAGGGTCACA
GCAGTGATCATGGTGATGCACTCGGTGCTGCTGGGTGGCGAGGAGCAGCGCTACCTCCTG
GAGGCCGCAGAGGAGCTGGGCCTGACCGATGGCTCCCTGGTCTTCCTGCCCTTCGACACG
ATCCACTACGCCTTGTCCCCAGGCCCGGAGGCCTTGGCCGCACTCGCCAACAGCTCCCAG
CTTCGCAGGGCCCACGATGCCGTGCTCACCCTCACGCGCCACTGTCCCTCTGAAGGCAGC
GTGCTGGACAGCCTGCGCAGGGCTCAAGAGCGCCGCGAGCTGCCCTCTGACCTCAATCTG
CAGCAGGTCTCCCCACTCTTTGGCACCATCTATGACGCGGTCTTCTTGCTGGCAAGGGGC
GTGGCAGAAGCGCGGGCTGCCGCAGGTGGCAGATGGGTGTCCGGAGCAGCTGTGGCCCGC
CACATCCGGGATGCGCAGGTCCCTGGCTTCTGCGGGGACCTAGGAGGAGACGAGGAGCCC
CCATTCGTGCTGCTAGACACGGACGCGGCGGGAGACCGGCTTTTTGCCACATACATGCTG
GATCCTGCCCGGGGCTCCTTCCTCTCCGCCGGTACCCGGATGCACTTCCCGCGTGGGGGA
TCAGCACCCGGACCTGACCCCTCGTGCTGGTTCGATCCAAACAACATCTGCGGTGGAGGA
CTGGAGCCGGGCCTCGTCTTTCTTGGCTTCCTCCTGGTGGTTGGGATGGGGCTGGCTGGG
GCCTTCCTGGCCCATTATGTGAGGCACCGGCTACTTCACATGCAAATGGTCTCCGGCCCC
AACAAGATCATCCTGACCGTGGACGACATCACCTTTCTCCACCCACATGGGGGCACCTCT
CGAAAGGTGGCCCAGGGGAGTCGATCAAGTCTGGGTGCCCGCAGCATGTCAGACATTCGC
AGCGGCCCCAGCCAACACTTGGACAGCCCCAACATTGGTGTCTATGAGGGAGACAGGGTT
TGGCTGAAGAAATTCCCAGGGGATCAGCACATAGCTATCCGCCCAGCAACCAAGACGGCC
TTCTCCAAGCTCCAGGAGCTCCGGCATGAGAACGTGGCCCTCTACCTGGGGCTTTTCCTG
GCTCGGGGAGCAGAAGGCCCTGCGGCCCTCTGGGAGGGCAACCTGGCTGTGGTCTCAGAG
CACTGCACGCGGGGCTCTCTTCAGGACCTCCTCGCTCAGAGAGAAATAAAGCTGGACTGG
ATGTTCAAGTCCTCCCTCCTGCTGGACCTTATCAAGGGAATAAGGTATCTGCACCATCGA
GGCGTGGCTCATGGGCGGCTGAAGTCACGGAACTGCATAGTGGATGGCAGATTCGTACTC
AAGATCACTGACCACGGCCACGGGAGACTGCTGGAAGCACAGAAGGTGCTACCGGAGCCT
CCCAGAGCGGAGGACCAGCTGTGGACAGCCCCGGAGCTGCTTAGGGACCCAGCCCTGGAG
CGCCGGGGAACGCTGGCCGGCGACGTCTTTAGCTTGGCCATCATCATGCAAGAAGTAGTG
TGCCGCAGTGCCCCTTATGCCATGCTGGAGCTCACTCCCGAGGAAGTGGTGCAGAGGGTG
CGGAGCCCCCCTCCACTGTGTCGGCCCTTGGTGTCCATGGACCAGGCACCTGTCGAGTGT
ATCCTCCTGATGAAGCAGTGCTGGGCAGAGCAGCCGGAACTTCGGCCCTCCATGGACCAC
ACCTTCGACCTGTTCAAGAACATCAACAAGGGCCGGAAGACGAACATCATTGACTCGATG
CTTCGGATGCTGGAGCAGTACTCTAGTAACCTGGAGGATCTGATCCGGGAGCGCACGGAG
GAGCTGGAGCTGGAAAAGCAGAAGACAGACCGGCTGCTTACACAGATGCTGCCTCCGTCT
GTGGCTGAGGCCTTGAAGACGGGGACACCAGTGGAGCCCGAGTACTTTGAGCAAGTGACA
CTGTACTTTAGTGACATTGTGGGCTTCACCACCATCTCTGCCATGAGTGAGCCCATTGAG
GTTGTGGACCTGCTCAACGATCTCTACACACTCTTTGATGCCATCATTGGTTCCCACGAT
GTCTACAAGGTGGAGACAATAGGGGACGCCTATATGGTGGCCTCGGGGCTGCCCCAGCGG
AATGGGCAGCGACACGCGGCAGAGATCGCCAACATGTCACTGGACATCCTCAGTGCCGTG
GGCACTTTCCGCATGCGCCATATGCCTGAGGTTCCCGTGCGCATCCGCATAGGCCTGCAC
TCGGGTCCATGCGTGGCAGGCGTGGTGGGCCTCACCATGCCGCGGTACTGCCTGTTTGGG
GACACGGTCAACACCGCCTCGCGCATGGAGTCCACCGGGCTGCCTTACCGCATCCACGTG
AACTTGAGCACTGTGGGGATTCTCCGTGCTCTGGACTCGGGCTACCAGGTGGAGCTGCGA
GGCCGCACGGAGCTGAAGGGCAAGGGCGCCGAGGACACTTTCTGGCTAGTGGGCAGACGC
GGCTTCAACAAGCCCATCCCCAAACCGCCTGACCTGCAACCGGGGTCCAGCAACCACGGC
ATCAGCCTGCAGGAGATCCCACCCGAGCGGCGACGGAAGCTGGAGAAGGCGCGGCCGGGC
CAGTTCTCTTGA
|
| Protein Properties |
|---|
| Number of Residues
| 1103 |
| Molecular Weight
| 120057.18 |
| Theoretical pI
| 7.45 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Retinal guanylyl cyclase 1
MTACARRAGGLPDPGLCGPAWWAPSLPRLPRALPRLPLLLLLLLLQPPALSAVFTVGVLG
PWACDPIFSRARPDLAARLAAARLNRDPGLAGGPRFEVALLPEPCRTPGSLGAVSSALAR
VSGLVGPVNPAACRPAELLAEEAGIALVPWGCPWTQAEGTTAPAVTPAADALYALLRAFG
WARVALVTAPQDLWVEAGRSLSTALRARGLPVASVTSMEPLDLSGAREALRKVRDGPRVT
AVIMVMHSVLLGGEEQRYLLEAAEELGLTDGSLVFLPFDTIHYALSPGPEALAALANSSQ
LRRAHDAVLTLTRHCPSEGSVLDSLRRAQERRELPSDLNLQQVSPLFGTIYDAVFLLARG
VAEARAAAGGRWVSGAAVARHIRDAQVPGFCGDLGGDEEPPFVLLDTDAAGDRLFATYML
DPARGSFLSAGTRMHFPRGGSAPGPDPSCWFDPNNICGGGLEPGLVFLGFLLVVGMGLAG
AFLAHYVRHRLLHMQMVSGPNKIILTVDDITFLHPHGGTSRKVAQGSRSSLGARSMSDIR
SGPSQHLDSPNIGVYEGDRVWLKKFPGDQHIAIRPATKTAFSKLQELRHENVALYLGLFL
ARGAEGPAALWEGNLAVVSEHCTRGSLQDLLAQREIKLDWMFKSSLLLDLIKGIRYLHHR
GVAHGRLKSRNCIVDGRFVLKITDHGHGRLLEAQKVLPEPPRAEDQLWTAPELLRDPALE
RRGTLAGDVFSLAIIMQEVVCRSAPYAMLELTPEEVVQRVRSPPPLCRPLVSMDQAPVEC
ILLMKQCWAEQPELRPSMDHTFDLFKNINKGRKTNIIDSMLRMLEQYSSNLEDLIRERTE
ELELEKQKTDRLLTQMLPPSVAEALKTGTPVEPEYFEQVTLYFSDIVGFTTISAMSEPIE
VVDLLNDLYTLFDAIIGSHDVYKVETIGDAYMVASGLPQRNGQRHAAEIANMSLDILSAV
GTFRMRHMPEVPVRIRIGLHSGPCVAGVVGLTMPRYCLFGDTVNTASRMESTGLPYRIHV
NLSTVGILRALDSGYQVELRGRTELKGKGAEDTFWLVGRRGFNKPIPKPPDLQPGSSNHG
ISLQEIPPERRRKLEKARPGQFS
|
| External Links |
|---|
| GenBank ID Protein
| 2695890 |
| UniProtKB/Swiss-Prot ID
| Q02846 |
| UniProtKB/Swiss-Prot Entry Name
| GUC2D_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AJ222657 |
| GeneCard ID
| GUCY2D |
| GenAtlas ID
| GUCY2D |
| HGNC ID
| HGNC:4689 |
| References |
|---|
| General References
| Not Available |

