| Identification |
|---|
| HMDB Protein ID
| CDBP00893 |
| Secondary Accession Numbers
| Not Available |
| Name
| DNA polymerase beta |
| Description
| Not Available |
| Synonyms
|
Not Available
|
| Gene Name
| POLB |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in DNA binding |
| Specific Function
| Repair polymerase that plays a key role in base-excision repair. Has 5'-deoxyribose-5-phosphate lyase (dRP lyase) activity that removes the 5' sugar phosphate and also acts as a DNA polymerase that adds one nucleotide to the 3' end of the arising single-nucleotide gap. Conducts 'gap-filling' DNA synthesis in a stepwise distributive fashion rather than in a processive fashion as for other DNA polymerases.
|
| GO Classification
|
| Biological Process |
| response to ethanol |
| DNA-dependent DNA replication |
| pyrimidine dimer repair |
| base-excision repair |
| cell death |
| Cellular Component |
| cytoplasm |
| nucleoplasm |
| spindle microtubule |
| Function |
| dna polymerase activity |
| dna-directed dna polymerase activity |
| nucleic acid binding |
| dna binding |
| binding |
| catalytic activity |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| nucleotidyltransferase activity |
| Molecular Function |
| metal ion binding |
| DNA-directed DNA polymerase activity |
| damaged DNA binding |
| microtubule binding |
| lyase activity |
| Process |
| metabolic process |
| macromolecule metabolic process |
| cellular macromolecule metabolic process |
| dna metabolic process |
| dna replication |
| dna repair |
|
| Cellular Location
|
- Nucleus
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Base excision repair | Not Available | 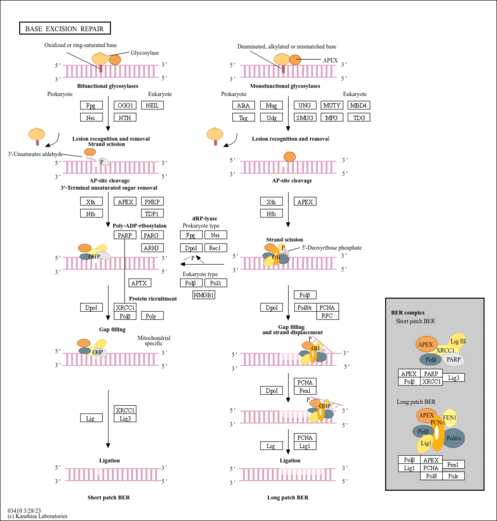 | | HTLV-I infection | Not Available |  | | Viral carcinogenesis | Not Available |  |
|
| Gene Properties |
|---|
| Chromosome Location
| 8 |
| Locus
| 8p11.2 |
| SNPs
| POLB |
| Gene Sequence
|
>1008 bp
ATGAGCAAACGGAAGGCGCCGCAGGAGACTCTCAACGGGGGAATCACCGACATGCTCACA
GAACTCGCAAACTTTGAGAAGAACGTGAGCCAAGCTATCCACAAGTACAATGCTTACAGA
AAAGCAGCATCTGTTATAGCAAAATACCCACACAAAATAAAGAGTGGAGCTGAAGCTAAG
AAATTGCCTGGAGTAGGAACAAAAATTGCTGAAAAGATTGATGAGTTTTTAGCAACTGGA
AAATTACGTAAACTGGAAAAGATTCGGCAGGATGATACGAGTTCATCCATCAATTTCCTG
ACTCGAGTTAGTGGCATTGGTCCATCTGCTGCAAGGAAGTTTGTAGATGAAGGAATTAAA
ACACTAGAAGATCTCAGAAAAAATGAAGATAAATTGAACCATCATCAGCGAATTGGGCTG
AAATATTTTGGGGACTTTGAAAAAAGAATTCCTCGTGAAGAGATGTTACAAATGCAAGAT
ATTGTACTAAATGAAGTTAAAAAAGTGGATTCTGAATACATTGCTACAGTCTGTGGCAGT
TTCAGAAGAGGTGCAGAGTCCAGTGGTGACATGGATGTTCTCCTGACCCATCCCAGCTTC
ACTTCAGAATCAACCAAACAGCCAAAACTGTTACATCAGGTTGTGGAGCAGTTACAAAAG
GTTCATTTTATCACAGATACCCTGTCAAAGGGTGAGACAAAGTTCATGGGTGTTTGCCAG
CTTCCCAGTAAAAATGATGAAAAAGAATATCCACACAGAAGAATTGATATCAGGTTGATA
CCCAAAGATCAGTATTACTGTGGTGTTCTCTATTTCACTGGGAGTGATATTTTCAATAAG
AATATGAGGGCTCATGCCCTAGAAAAGGGTTTCACAATCAATGAGTACACCATCCGTCCC
TTGGGAGTCACTGGAGTTGCAGGAGAACCCCTGCCAGTGGATAGTGAAAAAGACATCTTT
GATTACATCCAGTGGAAATACCGGGAACCCAAGGACCGGAGCGAATGA
|
| Protein Properties |
|---|
| Number of Residues
| 335 |
| Molecular Weight
| 38177.34 |
| Theoretical pI
| 8.947 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>DNA polymerase beta
MSKRKAPQETLNGGITDMLTELANFEKNVSQAIHKYNAYRKAASVIAKYPHKIKSGAEAK
KLPGVGTKIAEKIDEFLATGKLRKLEKIRQDDTSSSINFLTRVSGIGPSAARKFVDEGIK
TLEDLRKNEDKLNHHQRIGLKYFGDFEKRIPREEMLQMQDIVLNEVKKVDSEYIATVCGS
FRRGAESSGDMDVLLTHPSFTSESTKQPKLLHQVVEQLQKVHFITDTLSKGETKFMGVCQ
LPSKNDEKEYPHRRIDIRLIPKDQYYCGVLYFTGSDIFNKNMRAHALEKGFTINEYTIRP
LGVTGVAGEPLPVDSEKDIFDYIQWKYREPKDRSE
|
| External Links |
|---|
| GenBank ID Protein
| 292397 |
| UniProtKB/Swiss-Prot ID
| P06746 |
| UniProtKB/Swiss-Prot Entry Name
| DPOLB_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| L11607 |
| GeneCard ID
| POLB |
| GenAtlas ID
| POLB |
| HGNC ID
| HGNC:9174 |
| References |
|---|
| General References
| Not Available |


