| Identification |
|---|
| HMDB Protein ID
| CDBP00865 |
| Secondary Accession Numbers
| Not Available |
| Name
| D-3-phosphoglycerate dehydrogenase |
| Description
| Not Available |
| Synonyms
|
- 3-PGDH
|
| Gene Name
| PHGDH |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in phosphoglycerate dehydrogenase activity |
| Specific Function
| Not Available |
| GO Classification
|
| Biological Process |
| cellular nitrogen compound metabolic process |
| brain development |
| L-serine biosynthetic process |
| Cellular Component |
| cytosol |
| Function |
| nad or nadh binding |
| phosphoglycerate dehydrogenase activity |
| oxidoreductase activity, acting on the ch-oh group of donors, nad or nadp as acceptor |
| cofactor binding |
| oxidoreductase activity, acting on ch-oh group of donors |
| oxidoreductase activity |
| binding |
| nucleotide binding |
| catalytic activity |
| Molecular Function |
| electron carrier activity |
| NAD binding |
| phosphoglycerate dehydrogenase activity |
| Process |
| l-serine biosynthetic process |
| metabolic process |
| cellular metabolic process |
| serine family amino acid metabolic process |
| oxidation reduction |
| l-serine metabolic process |
| cellular amino acid and derivative metabolic process |
| cellular amino acid metabolic process |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Glycine and Serine Metabolism | 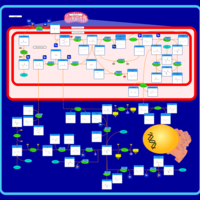 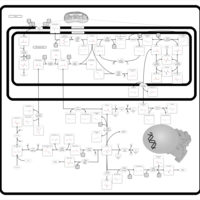 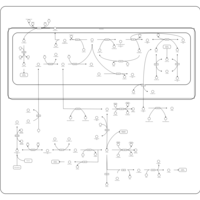 |  | | L-serine biosynthesis | Not Available | Not Available | | Glycine, serine and threonine metabolism | Not Available |  | | Dimethylglycine Dehydrogenase Deficiency | 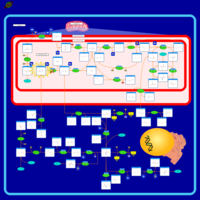 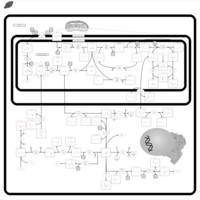  | Not Available | | Dihydropyrimidine Dehydrogenase Deficiency (DHPD) | 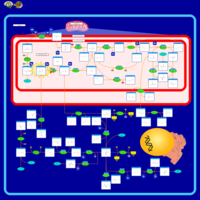 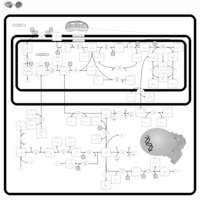 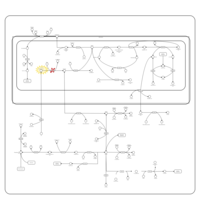 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 1 |
| Locus
| 1p12 |
| SNPs
| PHGDH |
| Gene Sequence
|
>1602 bp
ATGGCTTTTGCAAATCTGCGGAAAGTGCTCATCAGTGACAGCCTGGACCCTTGCTGCCGG
AAGATCTTGCAAGAGGGAGGGCTGCAGGTGGTGGAAAAGCAGAACCTTAGCAAAGAGGAG
CTGATAGCGGAGCTGCAGGACTGTGAAGGCCTTATTGTTCGCTCTGCCACCAAGGTGACC
GCTGATGTCATCAACGCAGCTGAGAAACTCCAGGTGGTGGGCAGGGCTGGCACAGGTGTG
GACAATGTGGATCTGGAGGCCGCAACAAGGAAGGGCATCTTGGTTATGAACACCCCCAAT
GGGAACAGCCTCAGTGCCGCAGAACTCACTTGTGGAATGATCATGTGCCTGGCCAGGCAG
ATTCCCCAGGCGACGGCTTCGATGAAGGACGGCAAATGGGAGCGGAAGAAGTTCATGGGA
ACAGAGCTGAATGGAAAGACCCTGGGAATTCTTGGCCTGGGCAGGATTGGGAGAGAGGTA
GCTACCCGGATGCAGTCCTTTGGGATGAAGACTATAGGGTATGACCCCATCATTTCCCCA
GAGGTCTCGGCCTCCTTTGGTGTTCAGCAGCTGCCCCTGGAGGAGATCTGGCCTCTCTGT
GATTTCATCACTGTGCACACTCCTCTCCTGCCCTCCACGACAGGCTTGCTGAATGACAAC
ACCTTTGCCCAGTGCAAGAAGGGGGTGCGTGTGGTGAACTGTGCCCGTGGAGGGATCGTG
GACGAAGGCGCCCTGCTCCGGGCCCTGCAGTCTGGCCAGTGTGCCGGGGCTGCACTGGAC
GTGTTTACGGAAGAGCCGCCACGGGACCGGGCCTTGGTGGACCATGAGAATGTCATCAGC
TGTCCCCACCTGGGTGCCAGCACCAAGGAGGCTCAGAGCCGCTGTGGGGAGGAAATTGCT
GTTCAGTTCGTGGACATGGTGAAGGGGAAATCTCTCACGGGGGTTGTGAATGCCCAGGCC
CTTACCAGTGCCTTCTCTCCACACACCAAGCCTTGGATTGGTCTGGCAGAAGCTCTGGGG
ACACTGATGCGAGCCTGGGCTGGGTCCCCCAAAGGGACCATCCAGGTGATAACACAGGGA
ACATCCCTGAAGAATGCTGGGAACTGCCTAAGCCCCGCAGTCATTGTCGGCCTCCTGAAA
GAGGCTTCCAAGCAGGCGGATGTGAACTTGGTGAACGCTAAGCTGCTGGTGAAAGAGGCT
GGCCTCAATGTCACCACCTCCCACAGCCCTGCTGCACCAGGGGAGCAAGGCTTCGGGGAA
TGCCTCCTGGCCGTGGCCCTGGCAGGCGCCCCTTACCAGGCTGTGGGCTTGGTCCAAGGC
ACTACACCTGTACTGCAGGGGCTCAATGGAGCTGTCTTCAGGCCAGAAGTGCCTCTCCGC
AGGGACCTGCCCCTGCTCCTATTCCGGACTCAGACCTCTGACCCTGCAATGCTGCCTACC
ATGATTGGCCTCCTGGCAGAGGCAGGCGTGCGGCTGCTGTCCTACCAGACTTCACTGGTG
TCAGATGGGGAGACCTGGCACGTCATGGGCATCTCCTCCTTGCTGCCCAGCCTGGAAGCG
TGGAAGCAGCATGTGACTGAAGCCTTCCAGTTCCACTTCTAA
|
| Protein Properties |
|---|
| Number of Residues
| 533 |
| Molecular Weight
| 56650.03 |
| Theoretical pI
| 6.715 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>D-3-phosphoglycerate dehydrogenase
MAFANLRKVLISDSLDPCCRKILQDGGLQVVEKQNLSKEELIAELQDCEGLIVRSATKVT
ADVINAAEKLQVVGRAGTGVDNVDLEAATRKGILVMNTPNGNSLSAAELTCGMIMCLARQ
IPQATASMKDGKWERKKFMGTELNGKTLGILGLGRIGREVATRMQSFGMKTIGYDPIISP
EVSASFGVQQLPLEEIWPLCDFITVHTPLLPSTTGLLNDNTFAQCKKGVRVVNCARGGIV
DEGALLRALQSGQCAGAALDVFTEEPPRDRALVDHENVISCPHLGASTKEAQSRCGEEIA
VQFVDMVKGKSLTGVVNAQALTSAFSPHTKPWIGLAEALGTLMRAWAGSPKGTIQVITQG
TSLKNAGNCLSPAVIVGLLKEASKQADVNLVNAKLLVKEAGLNVTTSHSPAAPGEQGFGE
CLLAVALAGAPYQAVGLVQGTTPVLQGLNGAVFRPEVPLRRDLPLLLFRTQTSDPAMLPT
MIGLLAEAGVRLLSYQTSLVSDGETWHVMGISSLLPSLEAWKQHVTEAFQFHF
|
| External Links |
|---|
| GenBank ID Protein
| 2674062 |
| UniProtKB/Swiss-Prot ID
| O43175 |
| UniProtKB/Swiss-Prot Entry Name
| SERA_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF006043 |
| GeneCard ID
| PHGDH |
| GenAtlas ID
| PHGDH |
| HGNC ID
| HGNC:8923 |
| References |
|---|
| General References
| Not Available |
