| Identification |
|---|
| HMDB Protein ID
| CDBP00814 |
| Secondary Accession Numbers
| Not Available |
| Name
| Beta-hexosaminidase subunit beta |
| Description
| Not Available |
| Synonyms
|
- Beta-N-acetylhexosaminidase subunit beta
- Beta-hexosaminidase subunit beta chain A
- Beta-hexosaminidase subunit beta chain B
- Cervical cancer proto-oncogene 7 protein
- HCC-7
- Hexosaminidase subunit B
- N-acetyl-beta-glucosaminidase subunit beta
|
| Gene Name
| HEXB |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in beta-N-acetylhexosaminidase activity |
| Specific Function
| Responsible for the degradation of GM2 gangliosides, and a variety of other molecules containing terminal N-acetyl hexosamines, in the brain and other tissues.
|
| GO Classification
|
| Biological Process |
| myelination |
| male courtship behavior |
| oligosaccharide catabolic process |
| sensory perception of sound |
| oogenesis |
| locomotory behavior |
| phospholipid biosynthetic process |
| neuromuscular process controlling balance |
| N-acetylglucosamine metabolic process |
| lipid storage |
| carbohydrate metabolic process |
| regulation of cell shape |
| glycosphingolipid metabolic process |
| phospholipid metabolic process |
| cell death |
| penetration of zona pellucida |
| chondroitin sulfate catabolic process |
| keratan sulfate catabolic process |
| lysosome organization |
| hyaluronan catabolic process |
| cellular protein metabolic process |
| cellular calcium ion homeostasis |
| skeletal system development |
| positive regulation of transcription from RNA polymerase II promoter |
| ganglioside catabolic process |
| astrocyte cell migration |
| Cellular Component |
| lysosomal lumen |
| acrosomal vesicle |
| membrane |
| Function |
| beta-n-acetylhexosaminidase activity |
| binding |
| catalytic activity |
| hydrolase activity |
| hydrolase activity, acting on glycosyl bonds |
| hydrolase activity, hydrolyzing o-glycosyl compounds |
| ion binding |
| cation binding |
| hexosaminidase activity |
| Molecular Function |
| cation binding |
| protein homodimerization activity |
| beta-N-acetylhexosaminidase activity |
| beta-N-acetylglucosaminidase activity |
| protein heterodimerization activity |
| Process |
| metabolic process |
| primary metabolic process |
| carbohydrate metabolic process |
|
| Cellular Location
|
- Lysosome
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Other glycan degradation | Not Available | 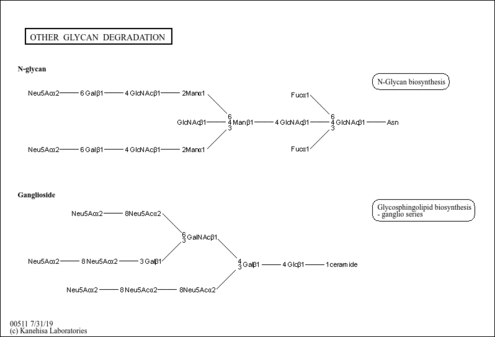 | | Amino sugar and nucleotide sugar metabolism | Not Available |  | | Glycosaminoglycan degradation | Not Available | 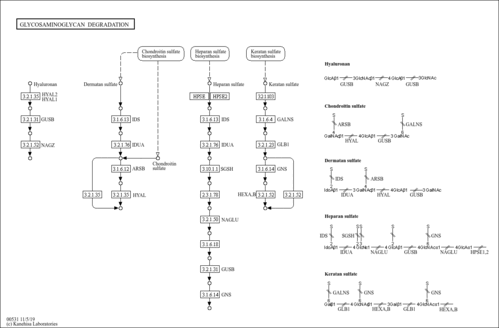 | | Glycosphingolipid biosynthesis - globo series | Not Available | 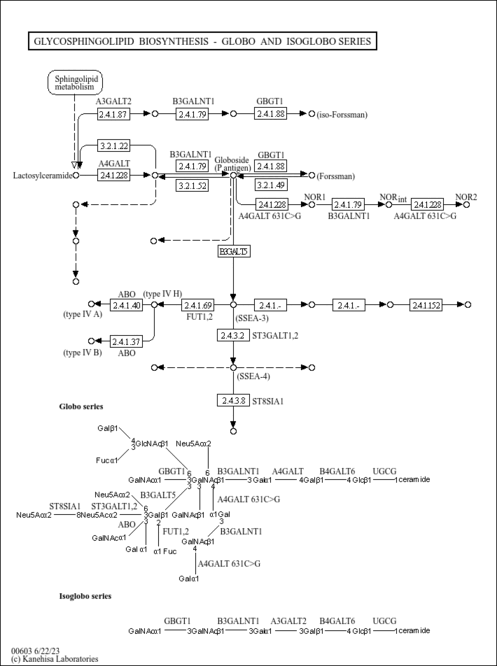 | | Glycosphingolipid biosynthesis - ganglio series | Not Available | 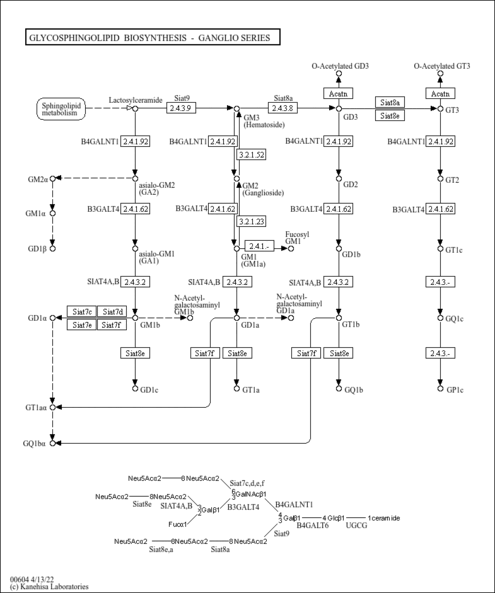 |
|
| Gene Properties |
|---|
| Chromosome Location
| Not Available |
| Locus
| Not Available |
| SNPs
| HEXB |
| Gene Sequence
|
>1671 bp
ATGGAGCTGTGCGGGCTGGGGCTGCCCCGGCCGCCCATGCTGCTGGCGCTGCTGTTGGCG
ACACTGCTGGCGGCGATGTTGGCGCTGCTGACTCAGGTGGCGCTGGTGGTGCAGGTGGCG
GAGGCGGCTCGGGCCCCGAGCGTCTCGGCCAAGCCGGGGCCGGCGCTGTGGCCCCTGCCG
CTCTCGGTGAAGATGACCCCGAACCTGCTGCATCTCGCCCCGGAGAACTTCTACATCAGC
CACAGCCCCAATTCCACGGCGGGCCCCTCCTGCACCCTGCTGGAGGAAGCGTTTCGACGA
TATCATGGCTATATTTTTGGTTTCTACAAGTGGCATCATGAACCTGCTGAATTCCAGGCT
AAAACCCAGGTTCAGCAACTTCTTGTCTCAATCACCCTTCAGTCAGAGTGTGATGCTTTC
CCCAACATATCTTCAGATGAGTCTTATACTTTACTTGTGAAAGAACCAGTGGCTGTCCTT
AAGGCCAACAGAGTTTGGGGAGCATTACGAGGTTTAGAGACCTTTAGCCAGTTAGTTTAT
CAAGATTCTTATGGAACTTTCACCATCAATGAATCCACCATTATTGATTCTCCAAGGTTT
TCTCACAGAGGAATTTTGATTGATACATCCAGACATTATCTGCCAGTTAAGATTATTCTT
AAAACTCTGGATGCCATGGCTTTTAATAAGTTTAATGTTCTTCACTGGCACATAGTTGAT
GACCAGTCTTTCCCATATCAGAGCATCACTTTTCCTGAGTTAAGCAATAAAGGAAGCTAT
TCTTTGTCTCATGTTTATACACCAAATGATGTCCGTATGGTGATTGAATATGCCAGATTA
CGAGGAATTCGAGTCCTGCCAGAATTTGATACCCCTGGGCATACACTATCTTGGGGAAAA
GGTCAGAAAGACCTCCTGACTCCATGTTACAGTAGACAAAACAAGTTGGACTCTTTTGGA
CCTATAAACCCTACTCTGAATACAACATACAGCTTCCTTACTACATTTTTCAAAGAAATT
AGTGAGGTGTTTCCAGATCAATTCATTCATTTGGGAGGAGATGAAGTGGAATTTAAATGT
TGGGAATCAAATCCAAAAATTCAAGATTTCATGAGGCAAAAAGGCTTTGGCACAGATTTT
AAGAAACTAGAATCTTTCTACATTCAAAAGGTTTTGGATATTATTGCAACCATAAACAAG
GGATCCATTGTCTGGCAGGAGGTTTTTGATGATAAAGCAAAGCTTGCGCCGGGCACAATA
GTTGAAGTATGGAAAGACAGCGCATATCCTGAGGAACTCAGTAGAGTCACAGCATCTGGC
TTCCCTGTAATCCTTTCTGCTCCTTGGTACTTAGATTTGATTAGCTATGGACAAGATTGG
AGGAAATACTATAAAGTGGAACCTCTTGATTTTGGCGGTACTCAGAAACAGAAACAACTT
TTCATTGGTGGAGAAGCTTGTCTATGGGGAGAATATGTGGATGCAACTAACCTCACTCCA
AGATTATGGCCTCGGGCAAGTGCTGTTGGTGAGAGACTCTGGAGTTCCAAAGATGTCAGA
GATATGGATGACGCCTATGACAGACTGACAAGGCACCGCTGCAGGATGGTCGAACGTGGA
ATAGCTGCACAACCTCTTTATGCTGGATATTGTAACCATGAGAACATGTAA
|
| Protein Properties |
|---|
| Number of Residues
| 556 |
| Molecular Weight
| Not Available |
| Theoretical pI
| Not Available |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Beta-hexosaminidase subunit beta
MELCGLGLPRPPMLLALLLATLLAAMLALLTQVALVVQVAEAARAPSVSAKPGPALWPLP
LSVKMTPNLLHLAPENFYISHSPNSTAGPSCTLLEEAFRRYHGYIFGFYKWHHEPAEFQA
KTQVQQLLVSITLQSECDAFPNISSDESYTLLVKEPVAVLKANRVWGALRGLETFSQLVY
QDSYGTFTINESTIIDSPRFSHRGILIDTSRHYLPVKIILKTLDAMAFNKFNVLHWHIVD
DQSFPYQSITFPELSNKGSYSLSHVYTPNDVRMVIEYARLRGIRVLPEFDTPGHTLSWGK
GQKDLLTPCYSRQNKLDSFGPINPTLNTTYSFLTTFFKEISEVFPDQFIHLGGDEVEFKC
WESNPKIQDFMRQKGFGTDFKKLESFYIQKVLDIIATINKGSIVWQEVFDDKAKLAPGTI
VEVWKDSAYPEELSRVTASGFPVILSAPWYLDLISYGQDWRKYYKVEPLDFGGTQKQKQL
FIGGEACLWGEYVDATNLTPRLWPRASAVGERLWSSKDVRDMDDAYDRLTRHRCRMVERG
IAAQPLYAGYCNHENM
|
| External Links |
|---|
| GenBank ID Protein
| 21309953 |
| UniProtKB/Swiss-Prot ID
| P07686 |
| UniProtKB/Swiss-Prot Entry Name
| HEXB_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF378118 |
| GeneCard ID
| HEXB |
| GenAtlas ID
| HEXB |
| HGNC ID
| HGNC:4879 |
| References |
|---|
| General References
| Not Available |




