| Identification |
|---|
| HMDB Protein ID
| CDBP00752 |
| Secondary Accession Numbers
| Not Available |
| Name
| Leukotriene C4 synthase |
| Description
| Not Available |
| Synonyms
|
- LTC4 synthase
- Leukotriene-C(4) synthase
|
| Gene Name
| LTC4S |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in enzyme activator activity |
| Specific Function
| Catalyzes the conjugation of leukotriene A4 with reduced glutathione to form leukotriene C4.
|
| GO Classification
|
| Biological Process |
| glutathione biosynthetic process |
| leukotriene biosynthetic process |
| response to axon injury |
| response to inorganic substance |
| response to drug |
| cellular response to lipopolysaccharide |
| cellular response to vitamin A |
| leukotriene metabolic process |
| positive regulation of catalytic activity |
| Cellular Component |
| endoplasmic reticulum membrane |
| endoplasmic reticulum |
| nuclear envelope |
| nuclear outer membrane |
| integral to membrane |
| Component |
| cell part |
| membrane part |
| intrinsic to membrane |
| integral to membrane |
| Function |
| enzyme regulator activity |
| enzyme activator activity |
| Molecular Function |
| glutathione transferase activity |
| lipid binding |
| enzyme activator activity |
| leukotriene-C4 synthase activity |
| glutathione peroxidase activity |
| glutathione binding |
| Process |
| metabolic process |
| cellular metabolic process |
| organic acid metabolic process |
| oxoacid metabolic process |
| carboxylic acid metabolic process |
| monocarboxylic acid metabolic process |
| fatty acid metabolic process |
| unsaturated fatty acid metabolic process |
| icosanoid metabolic process |
| leukotriene metabolic process |
|
| Cellular Location
|
- Endoplasmic reticulum membrane
- Multi-pass membrane protein
- Multi-pass membrane protein
- Nucleus outer membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Arachidonic Acid Metabolism |    |  | | Leukotriene C4 Synthesis Deficiency |    | Not Available | | Piroxicam Action Pathway |    | Not Available | | Acetylsalicylic Acid Action Pathway |  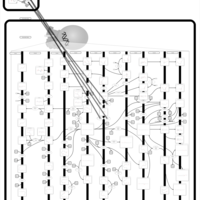 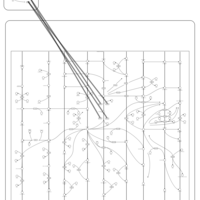 | Not Available | | Etodolac Action Pathway |  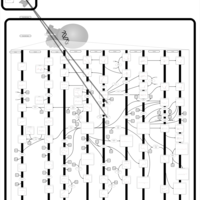  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 5 |
| Locus
| 5q35 |
| SNPs
| LTC4S |
| Gene Sequence
|
>453 bp
ATGAAGGACGAGGTAGCTCTACTGGCTGCTGTCACCCTCCTGGGAGTCCTGCTGCAAGCC
TACTTCTCCCTGCAGGTGATCTCGGCGCGCAGGGCCTTCCGCGTGTCGCCGCCGCTCACC
ACCGGCCCACCCGAGTTCGAGCGCGTCTACCGAGCCCAGGTGAACTGCAGCGAGTACTTC
CCGCTGTTCCTCGCCACGCTCTGGGTCGCCGGCATCTTCTTTCATGAAGGGGCGGCGGCC
CTGTGCGGCCTGGTCTACCTGTTCGCGCGCCTCCGCTACTTCCAGGGCTACGCGCGCTCC
GCGCAGCTCAGGCTGGCACCGCTGTACGCGAGCGCGCGCGCCCTCTGGCTGCTGGTGGCG
CTGGCTGCGCTCGGCCTGCTCGCCCACTTCCTCCCGGCCGCGCTGCGCGCCGCGCTCCTC
GGACGGCTCCGGACGCTGCTGCCGTGGGCCTGA
|
| Protein Properties |
|---|
| Number of Residues
| 150 |
| Molecular Weight
| 16566.465 |
| Theoretical pI
| 10.187 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Leukotriene C4 synthase
MKDEVALLAAVTLLGVLLQAYFSLQVISARRAFRVSPPLTTGPPEFERVYRAQVNCSEYF
PLFLATLWVAGIFFHEGAAALCGLVYLFARLRYFQGYARSAQLRLAPLYASARALWLLVA
LAALGLLAHFLPAALRAALLGRLRTLLPWA
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q16873 |
| UniProtKB/Swiss-Prot Entry Name
| LTC4S_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| U09353 |
| GeneCard ID
| LTC4S |
| GenAtlas ID
| LTC4S |
| HGNC ID
| HGNC:6719 |
| References |
|---|
| General References
| Not Available |
