| Identification |
|---|
| HMDB Protein ID
| CDBP00595 |
| Secondary Accession Numbers
| Not Available |
| Name
| Ribose-phosphate pyrophosphokinase 1 |
| Description
| Not Available |
| Synonyms
|
- PPRibP
- PRS-I
- Phosphoribosyl pyrophosphate synthase I
|
| Gene Name
| PRPS1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in magnesium ion binding |
| Specific Function
| Catalyzes the synthesis of phosphoribosylpyrophosphate (PRPP) that is essential for nucleotide synthesis.
|
| GO Classification
|
| Biological Process |
| carbohydrate metabolic process |
| pyrimidine nucleotide biosynthetic process |
| 5-phosphoribose 1-diphosphate biosynthetic process |
| hypoxanthine biosynthetic process |
| ribonucleoside monophosphate biosynthetic process |
| purine nucleotide biosynthetic process |
| nervous system development |
| nucleoside metabolic process |
| urate biosynthetic process |
| Cellular Component |
| cytosol |
| Function |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| diphosphotransferase activity |
| ribose phosphate diphosphokinase activity |
| magnesium ion binding |
| Molecular Function |
| ATP binding |
| magnesium ion binding |
| kinase activity |
| ribose phosphate diphosphokinase activity |
| Process |
| metabolic process |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside phosphate metabolic process |
| nucleotide metabolic process |
| nucleotide biosynthetic process |
| nucleoside monophosphate metabolic process |
| nucleoside monophosphate biosynthetic process |
| ribonucleoside monophosphate biosynthetic process |
| nucleoside metabolic process |
| biosynthetic process |
| cellular biosynthetic process |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Purine Metabolism |  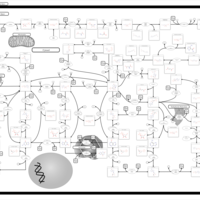 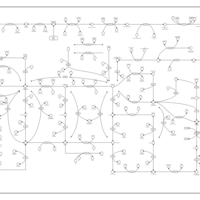 |  | | 5-phospho-alpha-D-ribose 1-diphosphate biosynthesis | Not Available | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| X |
| Locus
| Xq21.32-q24 |
| SNPs
| PRPS1 |
| Gene Sequence
|
>957 bp
ATGCCGAATATCAAAATCTTCAGCGGCAGCTCCCACCAGGACTTATCTCAGAAAATTGCT
GACCGCCTGGGCCTGGAGCTAGGCAAGGTGGTGACTAAGAAATTCAGCAACCAGGAGACC
TGTGTGGAAATTGGTGAAAGTGTACGTGGAGAGGATGTCTACATTGTTCAGAGTGGTTGT
GGCGAAATCAATGACAATTTAATGGAGCTTTTGATCATGATTAATGCCTGCAAGATTGCT
TCAGCCAGCCGGGTTACTGCAGTCATCCCATGCTTCCCTTATGCCCGGCAGGATAAGAAA
GATAAGAGCCGGGCGCCAATCTCAGCCAAGCTTGTTGCAAATATGCTATCTGTAGCAGGT
GCAGATCATATTATCACCATGGACCTACATGCTTCTCAAATTCAGGGCTTTTTTGATATC
CCAGTAGACAATTTGTATGCAGAGCCGGCTGTCCTAAAGTGGATAAGGGAGAATATCTCT
GAGTGGAGGAACTGCACTATTGTCTCACCTGATGCTGGTGGAGCTAAGAGAGTGACCTCC
ATTGCAGACAGGCTGAATGTGGACTTTGCCTTGATTCACAAAGAACGGAAGAAGGCCAAT
GAAGTGGACCGCATGGTGCTTGTGGGAGATGTGAAGGATCGGGTGGCCATCCTTGTGGAT
GACATGGCTGACACTTGTGGCACAATCTGCCATGCAGCTGACAAACTTCTCTCAGCTGGC
GCCACCAGAGTTTATGCCATCTTGACTCATGGAATCTTCTCCGGTCCTGCTATTTCTCGC
ATCAACAACGCATGCTTTGAGGCAGTAGTAGTCACCAATACCATACCTCAGGAGGACAAG
ATGAAGCATTGCTCCAAAATACAGGTGATTGACATCTCTATGATCCTTGCAGAAGCCATC
AGGAGAACTCACAATGGAGAATCCGTTTCTTACCTATTCAGCCATGTCCCTTTATAA
|
| Protein Properties |
|---|
| Number of Residues
| 318 |
| Molecular Weight
| 12324.195 |
| Theoretical pI
| 6.662 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Ribose-phosphate pyrophosphokinase 1
MPNIKIFSGSSHQDLSQKIADRLGLELGKVVTKKFSNQETCVEIGESVRGEDVYIVQSGC
GEINDNLMELLIMINACKIASASRVTAVIPCFPYARQDKKDKSRAPISAKLVANMLSVAG
ADHIITMDLHASQIQGFFDIPVDNLYAEPAVLKWIRENISEWRNCTIVSPDAGGAKRVTS
IADRLNVDFALIHKERKKANEVDRMVLVGDVKDRVAILVDDMADTCGTICHAADKLLSAG
ATRVYAILTHGIFSGPAISRINNACFEAVVVTNTIPQEDKMKHCSKIQVIDISMILAEAI
RRTHNGESVSYLFSHVPL
|
| External Links |
|---|
| GenBank ID Protein
| 57208781 |
| UniProtKB/Swiss-Prot ID
| P60891 |
| UniProtKB/Swiss-Prot Entry Name
| PRPS1_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AL137787 |
| GeneCard ID
| PRPS1 |
| GenAtlas ID
| PRPS1 |
| HGNC ID
| HGNC:9462 |
| References |
|---|
| General References
| Not Available |
