| Identification |
|---|
| HMDB Protein ID
| CDBP00577 |
| Secondary Accession Numbers
| Not Available |
| Name
| Ubiquitin-conjugating enzyme E2 D3 |
| Description
| Not Available |
| Synonyms
|
- Ubiquitin carrier protein D3
- Ubiquitin-conjugating enzyme E2(17)KB 3
- Ubiquitin-conjugating enzyme E2-17 kDa 3
- Ubiquitin-protein ligase D3
|
| Gene Name
| UBE2D3 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in acid-amino acid ligase activity |
| Specific Function
| Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins. In vitro catalyzes 'Lys-11'-, as well as 'Lys-48'-linked polyubiquitination. Cooperates with the E2 CDC34 and the SCF(FBXW11) E3 ligase complex for the polyubiquitination of NFKBIA leading to its subsequent proteasomal degradation. Acts as an initiator E2, priming the phosphorylated NFKBIA target at positions 'Lys-21' and/or 'Lys-22' with a monoubiquitin. Ubiquitin chain elongation is then performed by CDC34, building ubiquitin chains from the UBE2D3-primed NFKBIA-linked ubiquitin. Acts also as an initiator E2, in conjunction with RNF8, for the priming of PCNA. Monoubiquitination of PCNA, and its subsequent polyubiquitination, are essential events in the operation of the DNA damage tolerance (DDT) pathway that is activated after DNA damage caused by UV or chemical agents during S-phase. Associates with the BRCA1/BARD1 E3 ligase complex to perform ubiquitination at DNA damage sites following ionizing radiation leading to DNA repair. Targets DAPK3 for ubiquitination which influences promyelocytic leukemia protein nuclear body (PML-NB) formation in the nucleus. In conjunction with the MDM2 and TOPORS E3 ligases, functions ubiquitination of p53/TP53. Supports NRDP1-mediated ubiquitination and degradation of ERBB3 and of BRUCE which triggers apoptosis. In conjunction with the CBL E3 ligase, targets EGFR for polyubiquitination at the plasma membrane as well as during its internalization and transport on endosomes. In conjunction with the STUB1 E3 quality control E3 ligase, ubiquitinates unfolded proteins to catalyze their immediate destruction (By similarity).
|
| GO Classification
|
| Biological Process |
| protein K48-linked ubiquitination |
| protein K11-linked ubiquitination |
| protein monoubiquitination |
| negative regulation of type I interferon production |
| innate immune response |
| negative regulation of transcription from RNA polymerase II promoter |
| regulation of transcription from RNA polymerase II promoter in response to hypoxia |
| transforming growth factor beta receptor signaling pathway |
| proteasomal ubiquitin-dependent protein catabolic process |
| transcription initiation from RNA polymerase II promoter |
| apoptotic process |
| DNA repair |
| BMP signaling pathway |
| Cellular Component |
| cytosol |
| plasma membrane |
| nucleoplasm |
| endosome membrane |
| Function |
| ligase activity |
| ligase activity, forming carbon-nitrogen bonds |
| acid-amino acid ligase activity |
| catalytic activity |
| small conjugating protein ligase activity |
| Molecular Function |
| ubiquitin-protein ligase activity |
| ATP binding |
| Process |
| metabolic process |
| regulation of protein metabolic process |
| macromolecule metabolic process |
| biological regulation |
| regulation of biological process |
| regulation of metabolic process |
| regulation of macromolecule metabolic process |
| post-translational protein modification |
| macromolecule modification |
| protein modification process |
|
| Cellular Location
|
- Cell membrane
- Peripheral membrane protein
- Peripheral membrane protein
- Endosome membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | protein ubiquitination | Not Available | Not Available | | Ubiquitin mediated proteolysis | Not Available | 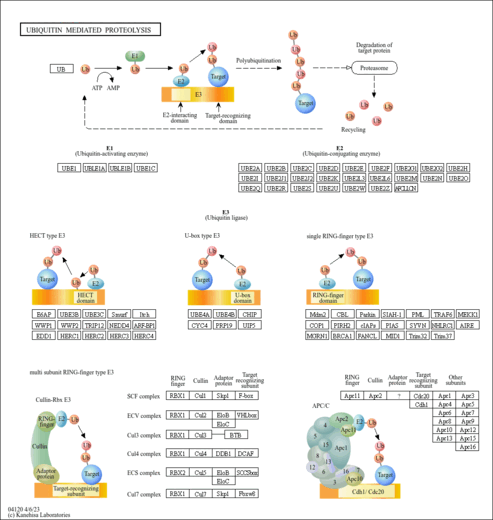 | | Protein processing in endoplasmic reticulum | Not Available |  |
|
| Gene Properties |
|---|
| Chromosome Location
| 4 |
| Locus
| 4q24 |
| SNPs
| UBE2D3 |
| Gene Sequence
|
>444 bp
ATGGCGCTGAAACGGATTAATAAGGAACTTAGTGATTTGGCCCGTGACCCTCCAGCACAA
TGTTCTGCAGGTCCAGTTGGGGATGATATGTTTCATTGGCAAGCCACAATTATGGGACCT
AATGACAGCCCATATCAAGGCGGTGTATTCTTTTTGACAATTCATTTTCCTACAGACTAC
CCCTTCAAACCACCTAAGGTTGCATTTACAACAAGAATTTATCATCCAAATATTAACAGT
AATGGCAGCATTTGTCTCGATATTCTAAGATCACAGTGGTCGCCTGCTTTAACAATTTCT
AAAGTTCTTTTATCCATTTGTTCACTGCTATGTGATCCAAACCCAGATGACCCCCTAGTG
CCAGAGATTGCACGGATCTATAAAACAGACAGAGATAAGTACAACAGAATATCTCGGGAA
TGGACTCAGAAGTATGCCATGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 147 |
| Molecular Weight
| 16686.96 |
| Theoretical pI
| 7.804 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Ubiquitin-conjugating enzyme E2 D3
MALKRINKELSDLARDPPAQCSAGPVGDDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDY
PFKPPKVAFTTRIYHPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLV
PEIARIYKTDRDKYNRISREWTQKYAM
|
| External Links |
|---|
| GenBank ID Protein
| 4507777 |
| UniProtKB/Swiss-Prot ID
| P61077 |
| UniProtKB/Swiss-Prot Entry Name
| UB2D3_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_003340.5 |
| GeneCard ID
| UBE2D3 |
| GenAtlas ID
| UBE2D3 |
| HGNC ID
| HGNC:12476 |
| References |
|---|
| General References
| Not Available |

