| Identification |
|---|
| HMDB Protein ID
| CDBP00556 |
| Secondary Accession Numbers
| Not Available |
| Name
| Ubiquitin-conjugating enzyme E2 D2 |
| Description
| Not Available |
| Synonyms
|
- Ubiquitin carrier protein D2
- Ubiquitin-conjugating enzyme E2(17)KB 2
- Ubiquitin-conjugating enzyme E2-17 kDa 2
- Ubiquitin-protein ligase D2
- p53-regulated ubiquitin-conjugating enzyme 1
|
| Gene Name
| UBE2D2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in acid-amino acid ligase activity |
| Specific Function
| Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins. In vitro catalyzes 'Lys-48'-linked polyubiquitination. Mediates the selective degradation of short-lived and abnormal proteins. Functions in the E6/E6-AP-induced ubiquitination of p53/TP53. Mediates ubiquitination of PEX5 and autoubiquitination of STUB1 and TRAF6. Involved in the signal-induced conjugation and subsequent degradation of NFKBIA, FBXW2-mediated GCM1 ubiquitination and degradation, MDM2-dependent degradation of p53/TP53 and the activation of MAVS in the mitochondria by DDX58/RIG-I in response to viral infection. Essential for viral activation of IRF3.
|
| GO Classification
|
| Biological Process |
| protein K48-linked ubiquitination |
| ubiquitin-dependent protein catabolic process |
| regulation of transcription from RNA polymerase II promoter in response to hypoxia |
| Cellular Component |
| cytosol |
| nucleoplasm |
| Function |
| catalytic activity |
| small conjugating protein ligase activity |
| ligase activity |
| ligase activity, forming carbon-nitrogen bonds |
| acid-amino acid ligase activity |
| Molecular Function |
| acid-amino acid ligase activity |
| ubiquitin-protein ligase activity |
| ATP binding |
| Process |
| metabolic process |
| regulation of protein metabolic process |
| macromolecule metabolic process |
| biological regulation |
| regulation of biological process |
| regulation of metabolic process |
| regulation of macromolecule metabolic process |
| post-translational protein modification |
| macromolecule modification |
| protein modification process |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | protein ubiquitination | Not Available | Not Available | | Ubiquitin mediated proteolysis | Not Available | 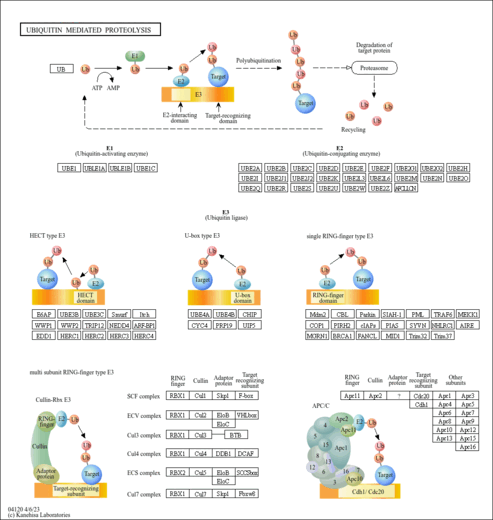 | | Protein processing in endoplasmic reticulum | Not Available | 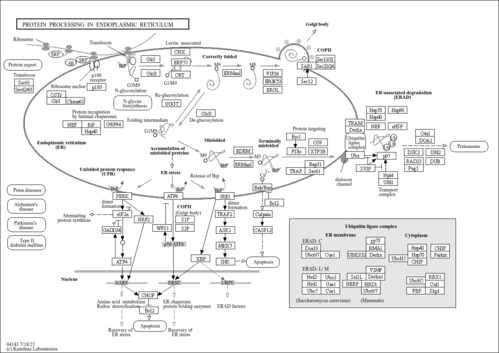 | | Shigellosis | Not Available | 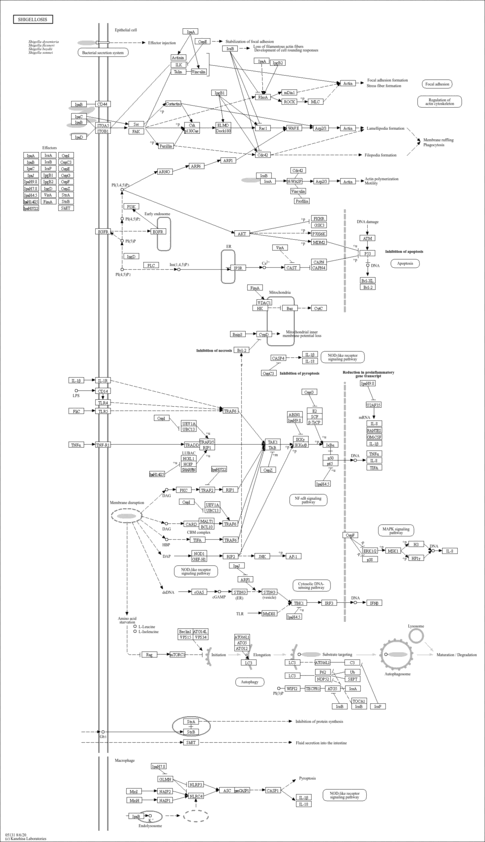 |
|
| Gene Properties |
|---|
| Chromosome Location
| 5 |
| Locus
| 5q31.2 |
| SNPs
| UBE2D2 |
| Gene Sequence
|
>444 bp
ATGGCTCTGAAGAGAATCCACAAGGAATTGAATGATCTGGCACGGGACCCTCCAGCACAG
TGTTCAGCAGGTCCTGTTGGAGATGATATGTTCCATTGGCAAGCTACAATAATGGGGCCA
AATGACAGTCCCTATCAGGGTGGAGTATTTTTCTTGACAATTCATTTCCCAACAGATTAC
CCCTTCAAACCACCTAAGGTTGCATTTACAACAAGAATTTATCATCCAAATATTAACAGT
AATGGCAGCATTTGTCTTGATATTCTACGATCACAGTGGTCTCCAGCACTAACTATTTCA
AAAGTACTCTTGTCCATCTGTTCTCTGTTGTGTGATCCCAATCCAGATGATCCTTTAGTG
CCTGAGATTGCTCGGATCTACAAAACAGATAGAGAAAAGTACAACAGAATAGCTCGGGAA
TGGACTCAGAAGTATGCGATGTAA
|
| Protein Properties |
|---|
| Number of Residues
| 147 |
| Molecular Weight
| 16735.06 |
| Theoretical pI
| 7.823 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Ubiquitin-conjugating enzyme E2 D2
MALKRIHKELNDLARDPPAQCSAGPVGDDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDY
PFKPPKVAFTTRIYHPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLV
PEIARIYKTDREKYNRIAREWTQKYAM
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| P62837 |
| UniProtKB/Swiss-Prot Entry Name
| UB2D2_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| U39317 |
| GeneCard ID
| UBE2D2 |
| GenAtlas ID
| UBE2D2 |
| HGNC ID
| HGNC:12475 |
| References |
|---|
| General References
| Not Available |


