| Identification |
|---|
| HMDB Protein ID
| CDBP00332 |
| Secondary Accession Numbers
| Not Available |
| Name
| Nicotinamide mononucleotide adenylyltransferase 2 |
| Description
| Not Available |
| Synonyms
|
- NMN adenylyltransferase 2
- NaMN adenylyltransferase 2
- Nicotinate-nucleotide adenylyltransferase 2
|
| Gene Name
| NMNAT2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in nucleotidyltransferase activity |
| Specific Function
| Catalyzes the formation of NAD(+) from nicotinamide mononucleotide (NMN) and ATP. Can also use the deamidated form; nicotinic acid mononucleotide (NaMN) as substrate but with a lower efficiency. Cannot use triazofurin monophosphate (TrMP) as substrate. Also catalyzes the reverse reaction, i.e. the pyrophosphorolytic cleavage of NAD(+). For the pyrophosphorolytic activity prefers NAD(+), NADH and NAAD as substrates and degrades nicotinic acid adenine dinucleotide phosphate (NHD) less effectively. Fails to cleave phosphorylated dinucleotides NADP(+), NADPH and NAADP(+).
|
| GO Classification
|
| Biological Process |
| NAD biosynthetic process |
| water-soluble vitamin metabolic process |
| Cellular Component |
| nucleus |
| trans-Golgi network |
| late endosome |
| Golgi membrane |
| Function |
| catalytic activity |
| transferase activity |
| transferase activity, transferring phosphorus-containing groups |
| nucleotidyltransferase activity |
| Molecular Function |
| nicotinate-nucleotide adenylyltransferase activity |
| ATP binding |
| nicotinamide-nucleotide adenylyltransferase activity |
| Process |
| metabolic process |
| coenzyme biosynthetic process |
| pyridine nucleotide biosynthetic process |
| nicotinamide nucleotide biosynthetic process |
| nad biosynthetic process |
| cellular metabolic process |
| biosynthetic process |
| cofactor metabolic process |
| coenzyme metabolic process |
|
| Cellular Location
|
- Cytoplasm
- Golgi apparatus
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | NAD(+) biosynthesis | Not Available | Not Available | | Nicotinate and Nicotinamide Metabolism | 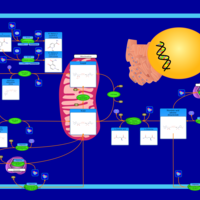 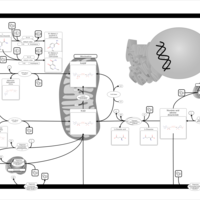  |  |
|
| Gene Properties |
|---|
| Chromosome Location
| 1 |
| Locus
| 1q25 |
| SNPs
| NMNAT2 |
| Gene Sequence
|
>924 bp
ATGACCGAGACCACCAAGACCCACGTTATCTTGCTCGCCTGCGGCAGCTTCAATCCCATC
ACCAAAGGGCACATTCAGATGTTTGAAAGAGCCAGGGATTATCTGCACAAAACTGGAAGG
TTTATTGTGATTGGCGGGATTGTCTCCCCTGTCCACGACTCCTATGGAAAACAGGGCCTC
GTGTCAAGCCGGCACCGTCTCATCATGTGTCAGCTGGCCGTCCAGAATTCTGATTGGATC
AGGGTGGACCCTTGGGAGTGCTACCAGGACACCTGGCAGACGACCTGCAGCGTGTTGGAA
CACCACCGGGACCTCATGAAGAGGGTGACTGGCTGCATCCTCTCCAATGTCAACACACCT
TCCATGACACCTGTGATCGGACAGCCACAAAACGAGACCCCCCAGCCCATTTACCAGAAC
AGCAACGTGGCCACCAAGCCCACTGCAGCCAAGATCTTGGGGAAGGTGGGAGAAAGCCTC
AGCCGGATCTGCTGTGTCCGCCCGCCGGTGGAGCGTTTCACCTTTGTAGATGAGAATGCC
AATCTGGGCACGGTGATGCGGTATGAAGAGATTGAGCTACGGATCCTGCTGCTGTGTGGT
AGTGACCTGCTGGAGTCCTTCTGCATCCCAGGGCTCTGGAACGAGGCAGATATGGAGGTG
ATTGTTGGTGACTTTGGGATTGTGGTGGTGCCCCGGGATGCAGCCGACACAGACCGAATC
ATGAATCACTCCTCAATACTCCGCAAATACAAAAACAACATCATGGTGGTGAAGGATGAC
ATCAACCATCCCATGTCTGTTGTCAGCTCAACCAAGAGCAGGCTGGCCCTGCAGCATGGG
GACGGCCATGTTGTGGATTACCTGTCCCAGCCGGTCATCGACTACATCCTCAAAAGCCAG
CTGTACATCAATGCCTCCGGCTAG
|
| Protein Properties |
|---|
| Number of Residues
| 307 |
| Molecular Weight
| 34438.38 |
| Theoretical pI
| 7.06 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Nicotinamide mononucleotide adenylyltransferase 2
MTETTKTHVILLACGSFNPITKGHIQMFERARDYLHKTGRFIVIGGIVSPVHDSYGKQGL
VSSRHRLIMCQLAVQNSDWIRVDPWECYQDTWQTTCSVLEHHRDLMKRVTGCILSNVNTP
SMTPVIGQPQNETPQPIYQNSNVATKPTAAKILGKVGESLSRICCVRPPVERFTFVDENA
NLGTVMRYEEIELRILLLCGSDLLESFCIPGLWNEADMEVIVGDFGIVVVPRDAADTDRI
MNHSSILRKYKNNIMVVKDDINHPMSVVSSTKSRLALQHGDGHVVDYLSQPVIDYILKSQ
LYINASG
|
| External Links |
|---|
| GenBank ID Protein
| 24307989 |
| UniProtKB/Swiss-Prot ID
| Q9BZQ4 |
| UniProtKB/Swiss-Prot Entry Name
| NMNA2_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| NM_015039.2 |
| GeneCard ID
| NMNAT2 |
| GenAtlas ID
| NMNAT2 |
| HGNC ID
| HGNC:16789 |
| References |
|---|
| General References
| Not Available |
