| Identification |
|---|
| HMDB Protein ID
| CDBP00082 |
| Secondary Accession Numbers
| Not Available |
| Name
| Soluble calcium-activated nucleotidase 1 |
| Description
| Not Available |
| Synonyms
|
- Apyrase homolog
- Putative MAPK-activating protein PM09
- Putative NF-kappa-B-activating protein 107
- SCAN-1
|
| Gene Name
| CANT1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in calcium ion binding |
| Specific Function
| Calcium-dependent nucleotidase with a preference for UDP. The order of activity with different substrates is UDP > GDP > UTP > GTP. Has very low activity towards ADP and even lower activity towards ATP. Does not hydrolyze AMP and GMP. Involved in proteoglycan synthesis.
|
| GO Classification
|
| Biological Process |
| positive regulation of I-kappaB kinase/NF-kappaB cascade |
| proteoglycan biosynthetic process |
| Cellular Component |
| endoplasmic reticulum membrane |
| Golgi cisterna membrane |
| integral to membrane |
| Function |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| hydrolase activity |
| calcium ion binding |
| hydrolase activity, acting on acid anhydrides |
| hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides |
| pyrophosphatase activity |
| Molecular Function |
| signal transducer activity |
| calcium ion binding |
| uridine-diphosphatase activity |
|
| Cellular Location
|
- Endoplasmic reticulum membrane
- Single-pass type II membrane protein
- Single-pass type II membrane protein
- Golgi apparatus
- Golgi stack membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Lactose Synthesis | 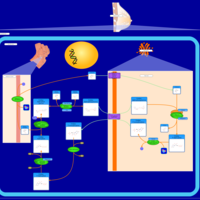 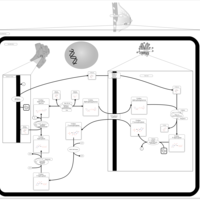 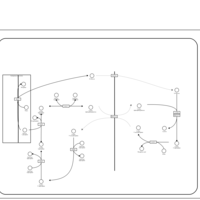 | Not Available | | Congenital disorder of glycosylation CDG-IId | 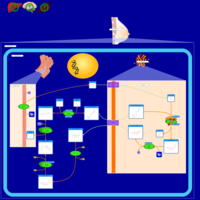 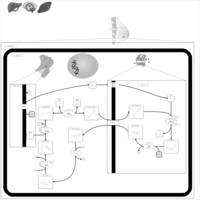 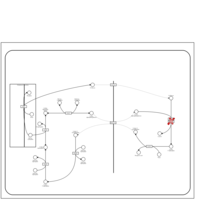 | Not Available | | GLUT-1 deficiency syndrome | 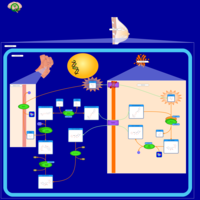 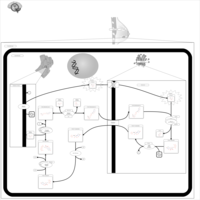 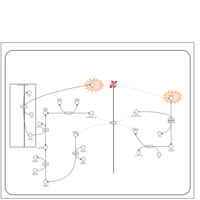 | Not Available | | Pyrimidine Metabolism |  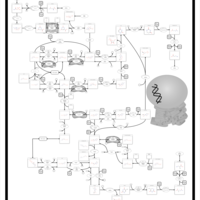  |  | | Beta Ureidopropionase Deficiency |  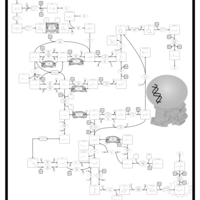 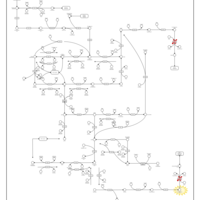 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 17 |
| Locus
| 17q25.3 |
| SNPs
| CANT1 |
| Gene Sequence
|
>1206 bp
ATGCCCGTGCAGCTGTCTGAGCACCCGGAATGGAATGAGTCTATGCACTCCCTCCGGATC
AGTGTGGGGGGCCTTCCTGTGCTGGCGTCCATGACCAAGGCCGCGGACCCCCGCTTCCGC
CCCCGCTGGAAGGTGATCCTGACGTTCTTTGTGGGTGCTGCCATCCTCTGGCTGCTCTGC
TCCCACCGCCCGGCCCCCGGCAGGCCCCCCACCCACAATGCACACAACTGGAGGCTCGGC
CAGGCGCCCGCCAACTGGTACAATGACACCTACCCCCTGTCTCCCCCACAAAGGACACCG
GCTGGGATTCGGTATCGAATCGCAGTTATCGCAGACCTGGACACAGAGTCAAGGGCCCAA
GAGGAAAACACCTGGTTCAGTTACCTGAAAAAGGGCTACCTGACCCTGTCAGACAGTGGG
GACAAGGTGGCCGTGGAATGGGACAAAGACCATGGGGTCCTGGAGTCCCACCTGGCGGAG
AAGGGGAGAGGCATGGAGCTATCCGACCTGATTGTTTTCAATGGGAAACTCTACTCCGTG
GATGACCGGACGGGGGTCGTCTACCAGATCGAAGGCAGCAAAGCCGTGCCCTGGGTGATT
CTGTCCGACGGCGACGGCACCGTGGAGAAAGGCTTCAAGGCCGAATGGCTGGCAGTGAAG
GACGAGCGTCTGTACGTGGGCGGCCTGGGCAAGGAGTGGACGACCACTACGGGTGATGTG
GTGAACGAGAACCCGGAGTGGGTGAAGGTGGTGGGCTACAAGGGCAGCGTGGACCACGAG
AACTGGGTGTCCAACTACAACGCCCTGCGGGCTGCTGCCGGCATCCAGCCGCCAGGCTAC
CTCATCCATGAGTCTGCCTGCTGGAGTGACACGCTGCAGCGCTGGTTCTTCCTGCCGCGC
CGCGCCAGCCAGGAGCGCTACAGCGAGAAGGACGACGAGCGCAAGGGCGCCAACCTGCTG
CTGAGCGCCTCCCCTGACTTCGGCGACATCGCTGTGAGCCACGTCGGGGCGGTGGTCCCC
ACTCACGGCTTCTCGTCCTTCAAGTTCATCCCCAACACCGACGACCAGATCATTGTGGCC
CTCAAATCCGAGGAGGACAGCGGCAGAGTCGCCTCCTACATCATGGCCTTCACGCTGGAC
GGGCGCTTCCTGTTGCCGGAGACCAAGATCGGAAGCGTGAAATACGAAGGCATCGAGTTC
ATTTAA
|
| Protein Properties |
|---|
| Number of Residues
| 401 |
| Molecular Weight
| 44839.24 |
| Theoretical pI
| 6.082 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Soluble calcium-activated nucleotidase 1
MPVQLSEHPEWNESMHSLRISVGGLPVLASMTKAADPRFRPRWKVILTFFVGAAILWLLC
SHRPAPGRPPTHNAHNWRLGQAPANWYNDTYPLSPPQRTPAGIRYRIAVIADLDTESRAQ
EENTWFSYLKKGYLTLSDSGDKVAVEWDKDHGVLESHLAEKGRGMELSDLIVFNGKLYSV
DDRTGVVYQIEGSKAVPWVILSDGDGTVEKGFKAEWLAVKDERLYVGGLGKEWTTTTGDV
VNENPEWVKVVGYKGSVDHENWVSNYNALRAAAGIQPPGYLIHESACWSDTLQRWFFLPR
RASQERYSEKDDERKGANLLLSASPDFGDIAVSHVGAVVPTHGFSSFKFIPNTDDQIIVA
LKSEEDSGRVASYIMAFTLDGRFLLPETKIGSVKYEGIEFI
|
| External Links |
|---|
| GenBank ID Protein
| 229577440 |
| UniProtKB/Swiss-Prot ID
| Q8WVQ1 |
| UniProtKB/Swiss-Prot Entry Name
| CANT1_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_001159772.1 |
| GeneCard ID
| CANT1 |
| GenAtlas ID
| CANT1 |
| HGNC ID
| HGNC:19721 |
| References |
|---|
| General References
| Not Available |
