| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:22:09 UTC |
|---|
| Updated at | 2021-01-04 20:37:47 UTC |
|---|
| CannabisDB ID | CDB006162 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Dimethylamine |
|---|
| Description | Dimethylamine, DMA, also known as (CH3)2NH, an organic secondary amine, belongs to the class of organic compounds known as dialkylamines. These are organic compounds containing a dialkylamine group, characterized by two alkyl groups bonded to the amino nitrogen. Dimethylamine is strong basic compound that has low toxicity. The DMA ammonium salt (CH3-NH2+-CH3) has a pKa of 10.73 which is higher than the pKa of methylamine (10.64) and trimethylamine (9.79). It is a colorless, liquefied and flammable gas with an ammonia and fish-like odor. It can also be found in solutions of up to 40% DMA in water. Dimethylamine is abundant in human urine, particularly after the consumption of fish and seafoods. The highest concentrations were obtained from individuals that consumed coley, squid and whiting with cod, haddock, sardine, skate and swordfish (PMID: 18282650 ). The main sources of urinary DMA include trimethylamine N-oxide, a common food component, and asymmetric dimethylarginine (ADMA), an endogenous inhibitor of nitric oxide (NO) synthesis. ADMA is excreted in the urine in part unmetabolized and in part after hydrolysis to DMA by dimethylarginine dimethylaminohydrolase (DDAH). In the body, DMA also undergoes nitrosation under weak acid conditions to give dimethlynitrosamine. DMA is an aminating agent in the manufacturing of ion-exchange resins that are used in food processing. DMA is a dehairing agent used in the in tanning process, in dyes, in rubber accelerators, in soaps and cleaning compounds and as an agricultural fungicide. Dimethylamine is produced by the catalytic reaction of methanol and ammonia at elevated temperatures and high pressure. Dimethylamine is a precursor to several industrially significant compounds. It reacts with carbon disulfide to give dimethyldithiocarbamate, a precursor to a family of chemicals widely used in the vulcanization of rubber. The solvents dimethylformamide and dimethylacetamide are derived from dimethylamine. It is a raw material used in the production of many agrichemicals and pharmaceuticals, such as dimefox and diphenhydramine, respectively. The chemical weapon tabun is derived from dimethylamine. The surfactant lauryl dimethylamine oxide is found in soaps and cleaning compounds. Unsymmetrical dimethylhydrazine, a rocket fuel, is prepared from dimethylamine. Dimethylamine reacts with acids to form salts, such as dimethylamine hydrochloride, an odorless white solid with a melting point of 171.5 °C. It is an attractant for boll weevils. Dimethylamine is also a constituent of cannabis smoke. It is volatilized during the combustion of cannabis ( Ref:DOI ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| DMA | ChEBI | | HNMe2 | ChEBI | | Me2nh | ChEBI | | N,N-Dimethylamine | ChEBI | | (CH3)2nh | Kegg | | Dimethylamine anhydrous | HMDB | | Dimethylamine anhydrous (dot) | HMDB | | Dimethylamine aqueous solution | HMDB | | Dimethylamine hydrobromide | HMDB | | Dimethylamine solution | HMDB | | N-Methyl-methanamine | HMDB | | N-Methylmethanamine | HMDB | | N-Methylmethanamine (acd/name 4.0) | HMDB | | Dimethylamine nitrate | HMDB | | Dimethylamine perchlorate | HMDB | | Dimethylamine sulfate | HMDB | | Dimethylamine hydrochloride | HMDB | | Dimethylamine phosphate (3:1) | HMDB | | Dimethylamine, conjugate acid | HMDB | | Dimethylammonium chloride | HMDB | | Dimethylamine sulfate (1:1) | HMDB | | Dimethylammonium formate | HMDB | | Dimethylamine monosulfate | HMDB |
|
|---|
| Chemical Formula | C2H7N |
|---|
| Average Molecular Weight | 45.08 |
|---|
| Monoisotopic Molecular Weight | 45.0578 |
|---|
| IUPAC Name | dimethylamine |
|---|
| Traditional Name | dimethylamine |
|---|
| CAS Registry Number | 124-40-3 |
|---|
| SMILES | CNC |
|---|
| InChI Identifier | InChI=1S/C2H7N/c1-3-2/h3H,1-2H3 |
|---|
| InChI Key | ROSDSFDQCJNGOL-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as dialkylamines. These are organic compounds containing a dialkylamine group, characterized by two alkyl groups bonded to the amino nitrogen. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Amines |
|---|
| Direct Parent | Dialkylamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Secondary aliphatic amine
- Organopnictogen compound
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Industrial application: Environmental role: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | -92.2 °C | Not Available | | Boiling Point | 7 - 9 °C | Wikipedia | | Water Solubility | 1630 mg/mL at 40 °C | Not Available | | logP | -0.38 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-0006-9000000000-8ae4e0d1cb5c91c730c0 | 2014-09-20 | View Spectrum | | GC-MS | Dimethylamine, non-derivatized, GC-MS Spectrum | splash10-0006-9000000000-cfb8e10b3b28eba99793 | Spectrum | | GC-MS | Dimethylamine, non-derivatized, GC-MS Spectrum | splash10-0006-9000000000-cfb8e10b3b28eba99793 | Spectrum | | Predicted GC-MS | Dimethylamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0002-9000000000-398c710ba4ea34342a91 | Spectrum | | Predicted GC-MS | Dimethylamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Dimethylamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80B) , Positive | splash10-0006-9000000000-8b085007dc085a460422 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0002-9000000000-103f1d373ec0d015b807 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-0002-9000000000-df28f146170e28bfcb5e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-0002-9000000000-cd82fdde4d83a74d83f1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-00wc-9100000000-aeb1521c62f5010e11aa | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-017i-9101000000-c269a0887eed5daf0e28 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0002-9000000000-103f1d373ec0d015b807 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0002-9000000000-df28f146170e28bfcb5e | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0002-9000000000-17646ed4f7c44ba14045 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-00wc-9100000000-aeb1521c62f5010e11aa | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-017i-9101000000-c269a0887eed5daf0e28 | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-933c4b104db02265ac4a | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-e85cf853d2e7110d18c1 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014j-9000000000-2032c41c4ca525ff4e1c | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-32c57cc32d61a2feae23 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-36fe99d2cc96e560f509 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-8914e111239eacc36328 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-46fb6b0f3d2dcd71d159 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-46fb6b0f3d2dcd71d159 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-46fb6b0f3d2dcd71d159 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-e97e92727205304cdb09 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-e97e92727205304cdb09 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-9cf36baccac69593c69f | 2021-09-23 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Citalopram Action Pathway | 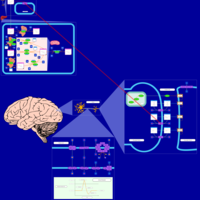 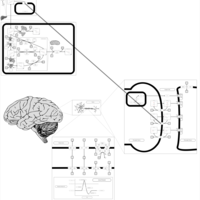  | Not Available | | Citalopram Metabolism Pathway | 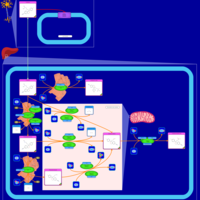   | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Amine oxidase [flavin-containing] B | MAOB | Xp11.23 | P27338 | details | | Amine oxidase [flavin-containing] A | MAOA | Xp11.3 | P21397 | details | | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | DDAH1 | 1p22 | O94760 | details | | N(G),N(G)-dimethylarginine dimethylaminohydrolase 2 | DDAH2 | 6p21.3 | O95865 | details |
|
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
| N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | DDAH1 | 1p22 | O94760 | details |
|
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000087 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012589 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 654 |
|---|
| KEGG Compound ID | C00543 |
|---|
| BioCyc ID | DIMETHYLAMINE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Dimethylamine |
|---|
| METLIN ID | 3758 |
|---|
| PubChem Compound | 674 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17170 |
|---|
| References |
|---|
| General References | - Mitchell SC, Zhang AQ, Smith RL: Dimethylamine and diet. Food Chem Toxicol. 2008 May;46(5):1734-8. doi: 10.1016/j.fct.2008.01.010. Epub 2008 Jan 15. [PubMed:18282650 ]
- Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, Argiles A: Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012 Jul;23(7):1258-70. doi: 10.1681/ASN.2011121175. Epub 2012 May 24. [PubMed:22626821 ]
- Meskys R, Harris RJ, Casaite V, Basran J, Scrutton NS: Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur J Biochem. 2001 Jun;268(12):3390-8. doi: 10.1046/j.1432-1327.2001.02239.x. [PubMed:11422368 ]
|
|---|