| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:29:15 UTC |
|---|
| Updated at | 2020-12-07 19:11:55 UTC |
|---|
| CannabisDB ID | CDB005299 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | 1-Methyluric acid |
|---|
| Description | 1-Methyluric acid, also known as 1-methylate, belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. An oxopurine that is 7,9-dihydro-1H-purine-2,6,8(3H)-trione substituted by a methyl group at N-1. 1-Methyluric acid is an extremely weak basic (essentially neutral) compound (based on its pKa). 1-Methyluric acid exists in all living organisms, ranging from bacteria to humans. 1-methyluric acid can be biosynthesized from 1-methylxanthine through the action of the enzyme xanthine dehydrogenase/oxidase. In humans, 1-methyluric acid is involved in caffeine metabolism. 1-Methyluric acid is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Methylate | Generator | | 1-Methylic acid | Generator | | 1-Methylurate | HMDB, MeSH |
|
|---|
| Chemical Formula | C6H6N4O3 |
|---|
| Average Molecular Weight | 182.14 |
|---|
| Monoisotopic Molecular Weight | 182.044 |
|---|
| IUPAC Name | 1-methyl-2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione |
|---|
| Traditional Name | 1-methyluric acid |
|---|
| CAS Registry Number | 708-79-2 |
|---|
| SMILES | CN1C(=O)NC2=C(NC(=O)N2)C1=O |
|---|
| InChI Identifier | InChI=1S/C6H6N4O3/c1-10-4(11)2-3(9-6(10)13)8-5(12)7-2/h1H3,(H,9,13)(H2,7,8,12) |
|---|
| InChI Key | QFDRTQONISXGJA-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Lactam
- Urea
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Source: Biological location: |
|---|
| Role | Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 5 mg/mL | Not Available | | logP | -0.57 | GASPARI,F & BONATI,M (1987) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | 1-Methyluric acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0far-2900000000-7c88ba8c892ca8263473 | Spectrum | | Predicted GC-MS | 1-Methyluric acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | 1-Methyluric acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-0900000000-f2e42cd67ce1106bed63 | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-0900000000-9fd2304504c5787dc255 | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a4i-9000000000-6069b457c0c038edb8df | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-0006-0900000000-f47eba015e7f8d9f21a7 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 4V, negative | splash10-001i-0900000000-26da10b11bacabae0d00 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 5V, negative | splash10-001i-0900000000-dceee0216914a6ad0349 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 6V, negative | splash10-001r-0900000000-cb9d81f2cc810b6a1bf6 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 8V, negative | splash10-0019-2900000000-79ddb513e243e4fd1054 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 10V, negative | splash10-001a-7900000000-aeb7c72625ab71bbfcb3 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 12V, negative | splash10-000i-0900000000-72f91c756d76768ecc23 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 12V, negative | splash10-004i-9000000000-0ae8d2338f48c17984cd | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 12V, negative | splash10-001i-9400000000-943dd572fd686e87913d | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-03di-0109000000-cf0f38f1f9c792f0404d | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 1V, negative | splash10-03di-0309000000-9a8aaf68c144b67fbc6e | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, negative | splash10-01q9-0905000000-d40f59a74d4b8cc4e1d6 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, negative | splash10-001i-0901000000-265af2cad1ab697d3249 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, negative | splash10-001i-0900000000-2b4e4dd83c0bad2269ae | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 4V, negative | splash10-001i-0900000000-c37839583f5e4e15cc46 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 9V, negative | splash10-001i-0900000000-6254efb7ddc081f6fd0c | 2020-07-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0900000000-9fb7b528d968cab88bd3 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0059-1900000000-eb2d1569b5bdfeef979a | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000w-9300000000-065aa3463c121bf31312 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-1900000000-62dd716e79c8fa6f5ee0 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-3900000000-bf85a8ad5b7719f36aba | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-7db60439b29417bbbe0d | 2017-09-01 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, 5%_DMSO, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 5%_DMSO, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Caffeine Metabolism | 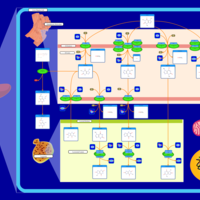 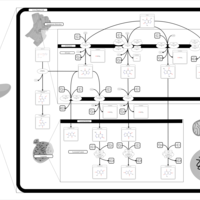  | 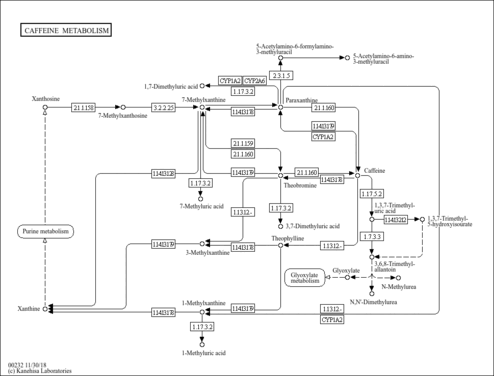 |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0003099 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB023107 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 62926 |
|---|
| KEGG Compound ID | C16359 |
|---|
| BioCyc ID | CPD-14119 |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| METLIN ID | 2821 |
|---|
| PubChem Compound | 69726 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 68441 |
|---|
| References |
|---|
| General References | Not Available |
|---|
