| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:27:16 UTC |
|---|
| Updated at | 2020-11-18 16:39:39 UTC |
|---|
| CannabisDB ID | CDB005279 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | 3,4-Dihydroxyphenylacetaldehyde |
|---|
| Description | 3,4-Dihydroxyphenylacetaldehyde, also known as protocatechuatealdehyde or 2-(3,4-dihydroxyphenyl)ethanal, belongs to the class of organic compounds known as phenylacetaldehydes. Phenylacetaldehydes are compounds containing a phenylacetaldehyde moiety, which consists of a phenyl group substituted at the second position by an acetalydehyde. 3,4-Dihydroxyphenylacetaldehyde is an extremely weak basic (essentially neutral) compound (based on its pKa). 3,4-Dihydroxyphenylacetaldehyde exists in all living organisms, ranging from bacteria to humans. Within humans, 3,4-dihydroxyphenylacetaldehyde participates in a number of enzymatic reactions. In particular, 3,4-dihydroxyphenylacetaldehyde can be biosynthesized from dopamine through its interaction with the enzyme amine oxidase [flavin-containing] a. In addition, 3,4-dihydroxyphenylacetaldehyde can be converted into 3,4-dihydroxybenzeneacetic acid through the action of the enzyme 3,4-dihydroxyphenylacetaldehyde. In humans, 3,4-dihydroxyphenylacetaldehyde is involved in the metabolic disorder called tyrosinemia type I. Outside of the human body, 3,4-Dihydroxyphenylacetaldehyde has been detected, but not quantified in, several different foods, such as pot marjorams, muscadine grapes, rambutans, summer savories, and cottonseeds. This could make 3,4-dihydroxyphenylacetaldehyde a potential biomarker for the consumption of these foods. A phenylacetaldehyde in which the 3 and 4 positions of the phenyl group are substituted by hydroxy groups. 3,4-Dihydroxyphenylacetaldehyde is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-(3,4-Dihydroxyphenyl)ethanal | ChEBI | | DOPAL | ChEBI | | Protocatechuatealdehyde | ChEBI | | DHPAA aldehyde | MeSH | | 3,4-Dihydroxy-benzeneacetaldehyde | HMDB |
|
|---|
| Chemical Formula | C8H8O3 |
|---|
| Average Molecular Weight | 152.15 |
|---|
| Monoisotopic Molecular Weight | 152.0473 |
|---|
| IUPAC Name | 2-(3,4-dihydroxyphenyl)acetaldehyde |
|---|
| Traditional Name | dopal |
|---|
| CAS Registry Number | 5707-55-1 |
|---|
| SMILES | OC1=C(O)C=C(CC=O)C=C1 |
|---|
| InChI Identifier | InChI=1S/C8H8O3/c9-4-3-6-1-2-7(10)8(11)5-6/h1-2,4-5,10-11H,3H2 |
|---|
| InChI Key | IADQVXRMSNIUEL-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as phenylacetaldehydes. Phenylacetaldehydes are compounds containing a phenylacetaldehyde moiety, which consists of a phenyl group substituted at the second position by an acetalydehyde. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Benzene and substituted derivatives |
|---|
| Sub Class | Phenylacetaldehydes |
|---|
| Direct Parent | Phenylacetaldehydes |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenylacetaldehyde
- Catechol
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Alpha-hydrogen aldehyde
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aldehyde
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | 1.005 | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | 3,4-Dihydroxyphenylacetaldehyde, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00di-3900000000-7f82d1d31cd24be9e865 | Spectrum | | Predicted GC-MS | 3,4-Dihydroxyphenylacetaldehyde, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00e9-8390000000-bba6c4376314951fb5ee | Spectrum | | Predicted GC-MS | 3,4-Dihydroxyphenylacetaldehyde, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-19827d81a06a62c2b70a | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udr-2900000000-479d55c57638faa6d9cd | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udl-9400000000-cadc5e95515b4cecf1f7 | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-b0ef024b4e5895ddea08 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-2900000000-352f5ee2c302c824867e | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9500000000-96da2068f1ed3e30ae47 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0pb9-0900000000-588f3b3d236131a4fa7a | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-3900000000-d3765e83b72ff4e9c933 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0zfr-9100000000-63064bac00f3091d89c3 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0900000000-b75a7231958cf1fc53ec | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0900000000-ed865ca228b5007b7ed4 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-9100000000-7cc4c0962b7233330b8c | 2021-09-22 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Tyrosine Metabolism | 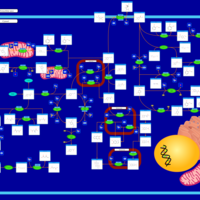 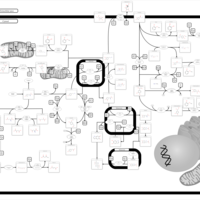 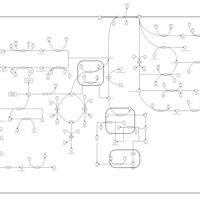 |  | | Alkaptonuria | 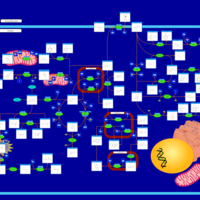 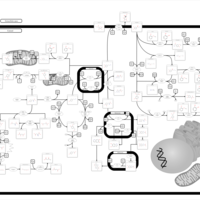 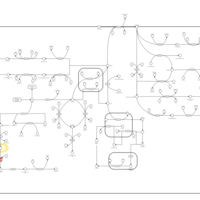 | Not Available | | Hawkinsinuria | 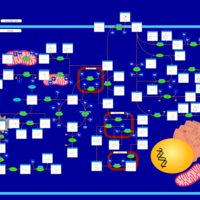  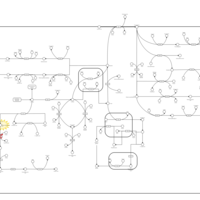 | Not Available | | Tyrosinemia Type I | 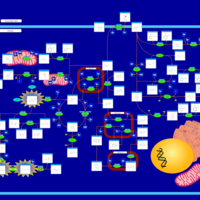 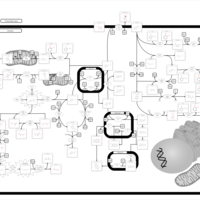 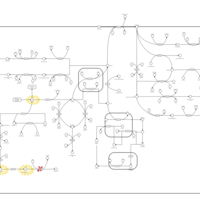 | Not Available | | Tyrosinemia, transient, of the newborn | 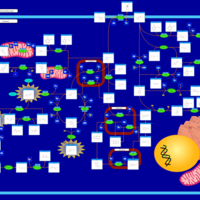 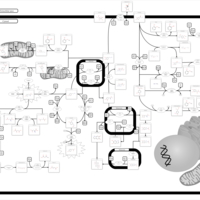 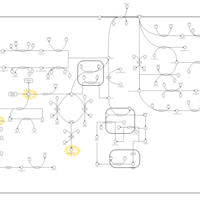 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Amine oxidase [flavin-containing] B | MAOB | Xp11.23 | P27338 | details | | Amine oxidase [flavin-containing] A | MAOA | Xp11.3 | P21397 | details | | Aldehyde dehydrogenase, dimeric NADP-preferring | ALDH3A1 | 17p11.2 | P30838 | details | | Aldehyde dehydrogenase family 1 member A3 | ALDH1A3 | 15q26.3 | P47895 | details | | Amiloride-sensitive amine oxidase [copper-containing] | ABP1 | 7q36.1 | P19801 | details | | Membrane primary amine oxidase | AOC3 | 17q21 | Q16853 | details | | Retina-specific copper amine oxidase | AOC2 | 17q21 | O75106 | details | | Aldehyde dehydrogenase family 3 member B2 | ALDH3B2 | 11q13 | P48448 | details | | Aldehyde dehydrogenase family 3 member B1 | ALDH3B1 | 11q13 | P43353 | details |
|
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
| Amiloride-sensitive amine oxidase [copper-containing] | ABP1 | 7q36.1 | P19801 | details |
|
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0003791 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB023227 |
|---|
| KNApSAcK ID | C00052036 |
|---|
| Chemspider ID | 106504 |
|---|
| KEGG Compound ID | C04043 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | 3,4-Dihydroxyphenylacetaldehyde |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 119219 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 27978 |
|---|
| References |
|---|
| General References | Not Available |
|---|
