| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:24:16 UTC |
|---|
| Updated at | 2020-11-18 16:39:36 UTC |
|---|
| CannabisDB ID | CDB005249 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Xylulose 5-phosphate |
|---|
| Description | Xylulose 5-phosphate, also known as xylulose-p or xu-5-p, belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. Xylulose 5-phosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). Xylulose 5-phosphate exists in all living species, ranging from bacteria to humans. Within humans, xylulose 5-phosphate participates in a number of enzymatic reactions. In particular, xylulose 5-phosphate can be biosynthesized from D-ribulose 5-phosphate; which is catalyzed by the enzyme ribulose-phosphate 3-epimerase. In addition, D-ribose 5-phosphate and xylulose 5-phosphate can be biosynthesized from D-glyceraldehyde 3-phosphate and D-sedoheptulose 7-phosphate through the action of the enzyme transketolase. In humans, xylulose 5-phosphate is involved in pentose phosphate pathway. Outside of the human body, Xylulose 5-phosphate has been detected, but not quantified in, several different foods, such as parsnips, cabbages, common beets, celery stalks, and ostrich ferns. This could make xylulose 5-phosphate a potential biomarker for the consumption of these foods. The D-enantiomer of xylulose 5-phosphate. Xylulose 5-phosphate is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5-O-Phosphono-D-threo-pentos-2-ulose | ChEBI | | 5-O-Phosphono-D-xylulose | ChEBI | | D-Xylulose-5-phosphate | ChEBI | | D-Xylulose-5-phosphoric acid | Generator | | Xylulose 5-phosphoric acid | Generator | | D-Xylulose 5-phosphate | HMDB | | D-Xylulose 5-PO4 | HMDB | | D-Xylulose-5-P | HMDB | | Xu-5-P | HMDB | | Xylulose-P | HMDB | | Xylulose-phosphate | HMDB | | Xylulose-5-phosphate | MeSH, HMDB | | Xylulose-5-phosphate, (D)-isomer | MeSH, HMDB | | Xylulose 5-phosphate | HMDB | | keto-D-Xylulose 5-phosphate | HMDB |
|
|---|
| Chemical Formula | C5H11O8P |
|---|
| Average Molecular Weight | 230.11 |
|---|
| Monoisotopic Molecular Weight | 230.0192 |
|---|
| IUPAC Name | {[(2R,3S)-2,3,5-trihydroxy-4-oxopentyl]oxy}phosphonic acid |
|---|
| Traditional Name | ribulose-5-phosphate |
|---|
| CAS Registry Number | 4212-65-1 |
|---|
| SMILES | OCC(=O)[C@@H](O)[C@H](O)COP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H11O8P/c6-1-3(7)5(9)4(8)2-13-14(10,11)12/h4-6,8-9H,1-2H2,(H2,10,11,12)/t4-,5-/m1/s1 |
|---|
| InChI Key | FNZLKVNUWIIPSJ-RFZPGFLSSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose phosphate
- Pentose-5-phosphate
- Monosaccharide phosphate
- Monoalkyl phosphate
- Acyloin
- Beta-hydroxy ketone
- Organic phosphoric acid derivative
- Alkyl phosphate
- Phosphoric acid ester
- Alpha-hydroxy ketone
- Ketone
- Secondary alcohol
- Polyol
- Hydrocarbon derivative
- Organic oxide
- Carbonyl group
- Alcohol
- Primary alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Xylulose 5-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0592-9800000000-d336b0e2eff1402939d8 | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, 3 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0a4i-9023200000-1dd7d8e1f64921beb234 | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_1_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_8, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_9, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_10, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_11, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_12, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_13, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_14, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Xylulose 5-phosphate, TMS_2_15, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - QTOF 4V, negative | splash10-004j-9170000000-997b74cb48ace33cccd6 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 23V, negative | splash10-004j-9000000000-6bc02f4fe0074c9abd68 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 25V, negative | splash10-004j-9000000000-bdadc167eb24a88aac33 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 27V, negative | splash10-004j-9000000000-93d31d83892e88720b87 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 30V, negative | splash10-004i-9000000000-10cd88e85162e6269751 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 33V, negative | splash10-004i-9000000000-e9f2bf02f0d01a879d20 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 35V, negative | splash10-004i-9000000000-732cd41f406b617c64a0 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 40V, negative | splash10-004i-9000000000-25a5607c50f3e9bb5a14 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 45V, negative | splash10-004i-9000000000-ea506b9429ed926c5f93 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 23V, negative | splash10-0002-9120000000-1172898b831a36b0afcb | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 25V, negative | splash10-0002-9110000000-132340cd62e8e4a0ccd2 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 27V, negative | splash10-0002-9110000000-297d4c6907686258e552 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 30V, negative | splash10-0002-9100000000-1cce49f995c912b1cc94 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 33V, negative | splash10-0002-9100000000-8bba2d7511cc9829eb36 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 35V, negative | splash10-0002-9100000000-89b3b4fdcb0326bfa0e0 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 40V, negative | splash10-0002-9100000000-91e850ff1cd255a2f7fd | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 45V, negative | splash10-002b-9100000000-9c43f84a30add3ebce50 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 16V, negative | splash10-0002-9000000000-75847d481a06c31fa479 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 16V, negative | splash10-004i-0091000000-0af17cfb2eb7eab94967 | 2020-07-21 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-3790000000-6f5913e9948fa7667e41 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0btd-9710000000-3724bb9f611bbdc1fe11 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9200000000-8bc9cecb2dddad8ca5e3 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002r-9520000000-c36fdbcc1d3b19b2fafd | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9100000000-b67b8ea484aa4ddb40e3 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-82b17cf4f5a8c927fe64 | 2015-09-15 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Pentose Phosphate Pathway | 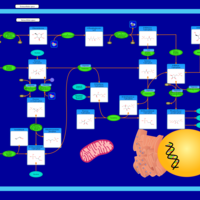  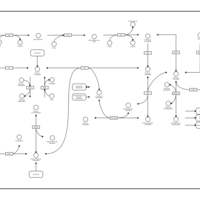 |  | | Glucose-6-phosphate dehydrogenase deficiency | 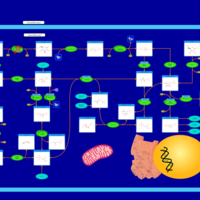 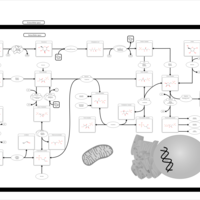 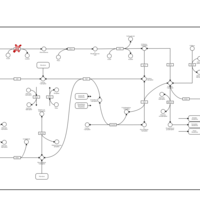 | Not Available | | Ribose-5-phosphate isomerase deficiency | 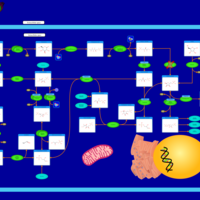 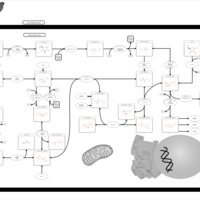 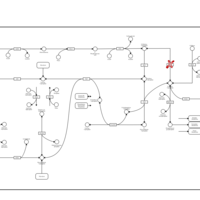 | Not Available | | Transaldolase deficiency | 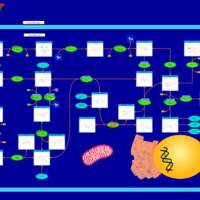 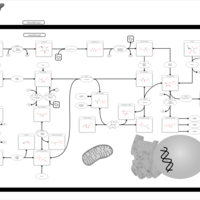 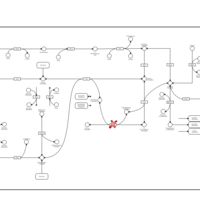 | Not Available | | Warburg Effect | 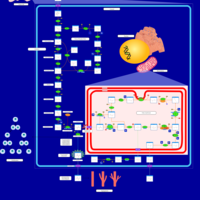 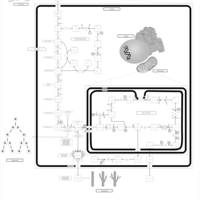 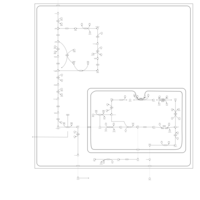 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000868 |
|---|
| DrugBank ID | DB04034 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB022290 |
|---|
| KNApSAcK ID | C00019693 |
|---|
| Chemspider ID | 388330 |
|---|
| KEGG Compound ID | C00231 |
|---|
| BioCyc ID | XYLULOSE-5-PHOSPHATE |
|---|
| BiGG ID | 34329 |
|---|
| Wikipedia Link | Xylulose 5-phosphate |
|---|
| METLIN ID | 5829 |
|---|
| PubChem Compound | 439190 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16332 |
|---|
| References |
|---|
| General References | Not Available |
|---|
