| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:51:58 UTC |
|---|
| Updated at | 2020-11-18 16:39:02 UTC |
|---|
| CannabisDB ID | CDB004933 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Ureidoisobutyric acid |
|---|
| Description | Ureidoisobutyric acid, also known as ureidoisobutyrate or beta-uba, belongs to the class of organic compounds known as ureas. Ureas are compounds containing two amine groups joined by a carbonyl (C=O) functional group. Ureidoisobutyric acid is an extremely weak basic (essentially neutral) compound (based on its pKa). Within humans, ureidoisobutyric acid participates in a number of enzymatic reactions. In particular, ureidoisobutyric acid can be biosynthesized from dihydrothymine; which is catalyzed by the enzyme dihydropyrimidinase. In addition, ureidoisobutyric acid can be converted into ureidoisobutyric acid; which is mediated by the enzyme Beta-ureidopropionase. In humans, ureidoisobutyric acid is involved in the metabolic disorder called the beta-ureidopropionase deficiency pathway. Ureidoisobutyric acid, with regard to humans, has been linked to the inborn metabolic disorder beta-ureidopropionase deficiency. Ureidoisobutyric acid is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Ureidoisobutyrate | Generator | | (2S)-3-[(C-Hydroxycarbonimidoyl)amino]-2-methylpropanoate | HMDB | | (2S)-3-(Carbamoylamino)-2-methylpropanoic acid | HMDB | | (2S)-3-(Carbamoylamino)-2-methylpropionic acid | HMDB | | 3-((Aminocarbonyl)amino)-2-methylpropanoic acid | HMDB | | 3-((Aminocarbonyl)amino)-2-methylpropionic acid | HMDB | | 3-Ureidoisobutyrate | HMDB | | 3-[(Aminocarbonyl)amino]-2-methylpropanoic acid | HMDB | | 3-[(Aminocarbonyl)amino]-2-methylpropionic acid | HMDB | | beta-UBA | HMDB | | beta-Ureidoisobutyric acid | HMDB | | Β-uba | HMDB | | Β-ureidoisobutyric acid | HMDB |
|
|---|
| Chemical Formula | C5H10N2O3 |
|---|
| Average Molecular Weight | 146.14 |
|---|
| Monoisotopic Molecular Weight | 146.0691 |
|---|
| IUPAC Name | (2S)-3-(carbamoylamino)-2-methylpropanoic acid |
|---|
| Traditional Name | (2S)-3-(carbamoylamino)-2-methylpropanoic acid |
|---|
| CAS Registry Number | 19140-82-0 |
|---|
| SMILES | C[C@@H](CNC(N)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H10N2O3/c1-3(4(8)9)2-7-5(6)10/h3H,2H2,1H3,(H,8,9)(H3,6,7,10)/t3-/m0/s1 |
|---|
| InChI Key | PHENTZNALBMCQD-VKHMYHEASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as ureas. Ureas are compounds containing two amine groups joined by a carbonyl (C=O) functional group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Organic carbonic acids and derivatives |
|---|
| Sub Class | Ureas |
|---|
| Direct Parent | Ureas |
|---|
| Alternative Parents | |
|---|
| Substituents | - Urea
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic nitrogen compound
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Ureidoisobutyric acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | Not Available | 2020-06-30 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | Not Available | 2020-06-30 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | Not Available | 2020-06-30 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | Not Available | 2020-06-30 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | Not Available | 2020-06-30 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | Not Available | 2020-06-30 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0019-8900000000-51286b64a41bad89fd80 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05gi-9100000000-835c9b02eecbd89baac8 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-40f2b4e3a071943603ee | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-3900000000-9dfc177176195ee47192 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052f-9000000000-0f56471f86b7f2230673 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-60a7db0ec3c9213e9347 | 2021-09-24 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Pyrimidine Metabolism |  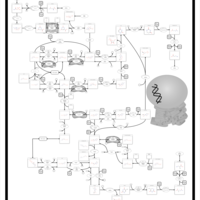  |  | | Beta Ureidopropionase Deficiency |  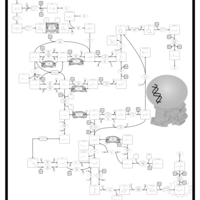 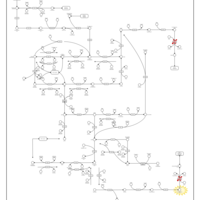 | Not Available | | UMP Synthase Deficiency (Orotic Aciduria) |  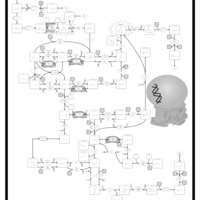  | Not Available | | Dihydropyrimidinase Deficiency |    | Not Available | | MNGIE (Mitochondrial Neurogastrointestinal Encephalopathy) |    | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0002031 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB022808 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C05100 |
|---|
| BioCyc ID | 3-UREIDO-ISOBUTYRATE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 88558720 |
|---|
| PDB ID | URQ |
|---|
| ChEBI ID | Not Available |
|---|
| References |
|---|
| General References | Not Available |
|---|
