| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:49:06 UTC |
|---|
| Updated at | 2020-11-18 16:38:58 UTC |
|---|
| CannabisDB ID | CDB004906 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | 6-Phosphogluconic acid |
|---|
| Description | 6-Phosphogluconic acid, also known as 6-phospho-D-gluconate or gluconate 6-phosphate, belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. A gluconic acid phosphate having the phosphate group at the 6-position. 6-Phosphogluconic acid is an extremely weak basic (essentially neutral) compound (based on its pKa). 6-Phosphogluconic acid exists in all living species, ranging from bacteria to humans. Within humans, 6-phosphogluconic acid participates in a number of enzymatic reactions. In particular, 6-phosphogluconic acid can be biosynthesized from gluconolactone through its interaction with the enzyme 6-phosphogluconolactonase. In addition, 6-phosphogluconic acid can be converted into D-ribulose 5-phosphate through the action of the enzyme 6-phosphogluconate dehydrogenase, decarboxylating. In humans, 6-phosphogluconic acid is involved in the metabolic disorder called the glucose-6-phosphate dehydrogenase deficiency pathway. Outside of the human body, 6-Phosphogluconic acid has been detected, but not quantified in, several different foods, such as teffs, chinese chestnuts, cottonseeds, irish moss, and red beetroots. This could make 6-phosphogluconic acid a potential biomarker for the consumption of these foods. 6-Phosphogluconic acid is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 6-Phospho-D-gluconate | ChEBI | | 6-Phospho-D-gluconic acid | Generator | | 6-Phosphogluconate | Generator | | 6-O-Phosphono-D-gluconic acid | HMDB | | 6-p-Gluconate | HMDB | | D-Gluconic acid 6-(dihydrogen phosphate) | HMDB | | D-Gluconic acid 6-phosphate | HMDB | | Gluconic acid-6-phosphate | HMDB | | Gluconate 6-phosphate | HMDB |
|
|---|
| Chemical Formula | C6H13O10P |
|---|
| Average Molecular Weight | 276.14 |
|---|
| Monoisotopic Molecular Weight | 276.0246 |
|---|
| IUPAC Name | (2R,3S,4R,5R)-2,3,4,5-tetrahydroxy-6-(phosphonooxy)hexanoic acid |
|---|
| Traditional Name | 6-phosphogluconic acid |
|---|
| CAS Registry Number | 921-62-0 |
|---|
| SMILES | O[C@H](COP(O)(O)=O)[C@@H](O)[C@H](O)[C@@H](O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H13O10P/c7-2(1-16-17(13,14)15)3(8)4(9)5(10)6(11)12/h2-5,7-10H,1H2,(H,11,12)(H2,13,14,15)/t2-,3-,4+,5-/m1/s1 |
|---|
| InChI Key | BIRSGZKFKXLSJQ-SQOUGZDYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Monosaccharide phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Gluconic_acid
- Hexose monosaccharide
- Monosaccharide phosphate
- Medium-chain hydroxy acid
- Medium-chain fatty acid
- Monoalkyl phosphate
- Beta-hydroxy acid
- Hydroxy fatty acid
- Alkyl phosphate
- Fatty acid
- Alpha-hydroxy acid
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Fatty acyl
- Hydroxy acid
- Secondary alcohol
- Polyol
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Carboxylic acid
- Carbonyl group
- Alcohol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | 6-Phosphogluconic acid, 5 TMS, GC-MS Spectrum | splash10-0uxu-0933000000-d0c837bb667abbae2ce0 | Spectrum | | GC-MS | 6-Phosphogluconic acid, 7 TMS, GC-MS Spectrum | splash10-00ks-1978000000-c7702d284ad9f06bb5c0 | Spectrum | | GC-MS | 6-Phosphogluconic acid, non-derivatized, GC-MS Spectrum | splash10-0uxu-0933000000-d0c837bb667abbae2ce0 | Spectrum | | GC-MS | 6-Phosphogluconic acid, non-derivatized, GC-MS Spectrum | splash10-00ks-1978000000-c7702d284ad9f06bb5c0 | Spectrum | | Predicted GC-MS | 6-Phosphogluconic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0a4j-9430000000-e4198ba354325082e116 | Spectrum | | Predicted GC-MS | 6-Phosphogluconic acid, 5 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00dl-7192047000-e8fa0bd25a22fa6fd28b | Spectrum | | Predicted GC-MS | 6-Phosphogluconic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | 6-Phosphogluconic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-00dj-5090000000-a1b0f01abec0e4fb22c5 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-002b-9000000000-780e6dc35ed33dde8b62 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-004i-9000000000-df8e5979c41c127eff41 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00b9-0495110000-a9d468f24ea04c2dd0d2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-9100000000-25ef4bb1cdb69b6418ce | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4j-5090000000-6db5e52f38f29e35f42a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0090000000-b6ed9e59bd4492f4b5ce | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-004i-0190000000-50702b7e8f4b7289d218 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-004j-9260000000-e356e1c75aa70d0065f9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-002b-9100000000-5be967e173f74d688dc2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-004j-9000000000-1c5eda88c214ef66d7b6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-004i-9000000000-e1b81e634eb90448b9cd | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-0090000000-50702b7e8f4b7289d218 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004j-9260000000-20df41bde9cbb406f532 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-002b-9100000000-56434d8c214a22e12603 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004j-9000000000-1c5eda88c214ef66d7b6 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-9000000000-e1b81e634eb90448b9cd | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0002-9100000000-25ef4bb1cdb69b6418ce | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0a4j-5090000000-6db5e52f38f29e35f42a | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-004i-0090000000-b6ed9e59bd4492f4b5ce | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-002b-9100000000-15a83e588180be4c0664 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-3d9937a328c6aa73fe8a | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-004j-7190000000-26e8b7b5ea3ee85cbb61 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-002b-9100000000-39b6ad01fb7aa0ecbb1a | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 19V, positive | splash10-01ow-3390000000-1a67f265724b65046d12 | 2020-07-22 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Pentose Phosphate Pathway | 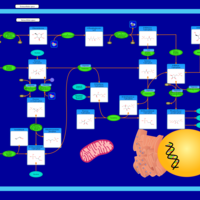  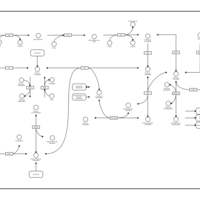 |  | | Glucose-6-phosphate dehydrogenase deficiency | 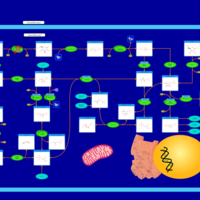 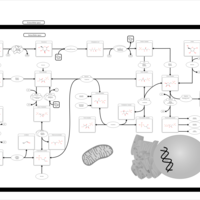 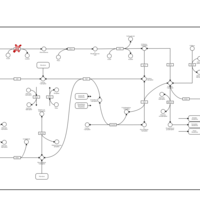 | Not Available | | Ribose-5-phosphate isomerase deficiency | 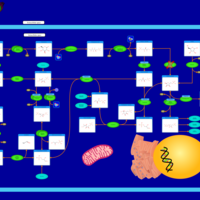 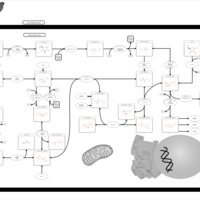 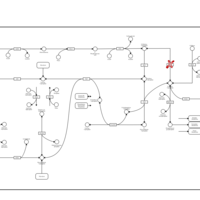 | Not Available | | Transaldolase deficiency | 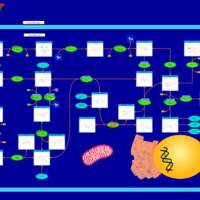 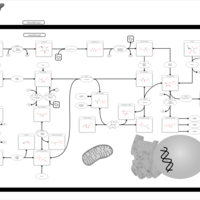 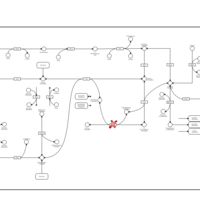 | Not Available | | Warburg Effect | 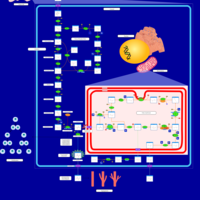 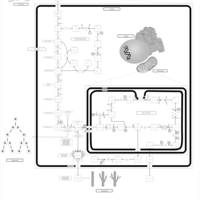 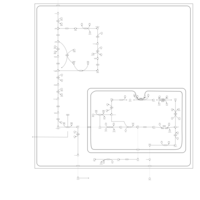 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | Not Available |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001316 |
|---|
| DrugBank ID | DB02076 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB030611 |
|---|
| KNApSAcK ID | C00007481 |
|---|
| Chemspider ID | 82615 |
|---|
| KEGG Compound ID | C00345 |
|---|
| BioCyc ID | CPD-2961 |
|---|
| BiGG ID | 34686 |
|---|
| Wikipedia Link | 6-Phosphogluconic_acid |
|---|
| METLIN ID | 367 |
|---|
| PubChem Compound | 91493 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 48928 |
|---|
| References |
|---|
| General References | Not Available |
|---|
