| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:37:33 UTC |
|---|
| Updated at | 2022-12-13 23:36:28 UTC |
|---|
| CannabisDB ID | CDB004795 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | cis-Aconitic acid |
|---|
| Description | cis-Aconitic acid, also known as cis-aconitate or acid, acontic, belongs to the class of organic compounds known as tricarboxylic acids and derivatives. These are carboxylic acids containing exactly three carboxyl groups. cis-Aconitic acid is a moderately acidic compound (based on its pKa). cis-Aconitic acid exists in all living species, ranging from bacteria to humans. Within humans, cis-aconitic acid participates in a number of enzymatic reactions. In particular, cis-aconitic acid can be biosynthesized from citric acid through its interaction with the enzyme aconitate hydratase, mitochondrial. In addition, cis-aconitic acid can be converted into isocitric acid through the action of the enzyme aconitate hydratase, mitochondrial. In humans, cis-aconitic acid is involved in the metabolic disorder called the glutaminolysis and cancer pathway. cis-Aconitic acid is a bland tasting compound. Outside of the human body, cis-Aconitic acid is found, on average, in the highest concentration within milk (cow). cis-Aconitic acid has also been detected, but not quantified in, several different foods, such as japanese pumpkins, purple mangosteens, opium poppies, root vegetables, and shallots. This could make cis-aconitic acid a potential biomarker for the consumption of these foods. The cis-isomer of aconitic acid. cis-Aconitic acid is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (Z)-1-Propene-1,2,3-tricarboxylic acid | ChEBI | | cis-1-Propene-1,2,3-tricarboxylic acid | ChEBI | | (Z)-1-Propene-1,2,3-tricarboxylate | Generator | | cis-1-Propene-1,2,3-tricarboxylate | Generator | | cis-Aconitate | Generator | | (1Z)-1-Propene-1,2,3-tricarboxylate | HMDB | | (1Z)-1-Propene-1,2,3-tricarboxylic acid | HMDB | | (Z)-Aconitate | HMDB | | (Z)-Aconitic acid | HMDB | | 1-cis-2,3-Propenetricarboxylate | HMDB | | 1-cis-2,3-Propenetricarboxylic acid | HMDB | | 1-Propene-1,2,3-tricarboxylate | HMDB | | 1-Propene-1,2,3-tricarboxylic acid | HMDB | | cis-Aconate | HMDB | | cis-Aconic acid | HMDB | | cis-Oxaloacetate | HMDB | | cis-Oxaloacetic acid | HMDB | | Acid, citridic | HMDB | | Acid, citridinic | HMDB | | Aconitic acid | HMDB | | Acontic acid | HMDB | | Adonic acid | HMDB | | Achilleic acid | HMDB | | Acid, acontic | HMDB | | Acid, equisetic | HMDB | | Citridinic acid | HMDB | | Acid, achilleic | HMDB | | Aconitate | HMDB | | Citridic acid | HMDB | | Pyrocitric acid | HMDB | | Acid, aconitic | HMDB | | Acid, adonic | HMDB | | Acid, carboxyglutaconic | HMDB | | Acid, pyrocitric | HMDB | | Carboxyglutaconic acid | HMDB | | Equisetic acid | HMDB |
|
|---|
| Chemical Formula | C6H6O6 |
|---|
| Average Molecular Weight | 174.11 |
|---|
| Monoisotopic Molecular Weight | 174.0164 |
|---|
| IUPAC Name | (1Z)-prop-1-ene-1,2,3-tricarboxylic acid |
|---|
| Traditional Name | cis-aconitic acid |
|---|
| CAS Registry Number | 585-84-2 |
|---|
| SMILES | OC(=O)C\C(=C\C(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H6O6/c7-4(8)1-3(6(11)12)2-5(9)10/h1H,2H2,(H,7,8)(H,9,10)(H,11,12)/b3-1- |
|---|
| InChI Key | GTZCVFVGUGFEME-IWQZZHSRSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as tricarboxylic acids and derivatives. These are carboxylic acids containing exactly three carboxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Tricarboxylic acids and derivatives |
|---|
| Direct Parent | Tricarboxylic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Tricarboxylic acid or derivatives
- Carboxylic acid
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 125 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 400 mg/mL | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | cis-Aconitic acid, 3 TMS, GC-MS Spectrum | splash10-0002-1940000000-24c70788b42e3006de46 | Spectrum | | GC-MS | cis-Aconitic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0940000000-3ab0a8161f2ec8f04452 | Spectrum | | GC-MS | cis-Aconitic acid, 3 TMS, GC-MS Spectrum | splash10-00dj-9630000000-1dc322333cb48ef39588 | Spectrum | | GC-MS | cis-Aconitic acid, 3 TMS, GC-MS Spectrum | splash10-004i-3591000000-a58ec1ba633b098e0867 | Spectrum | | GC-MS | cis-Aconitic acid, non-derivatized, GC-MS Spectrum | splash10-0002-1940000000-24c70788b42e3006de46 | Spectrum | | GC-MS | cis-Aconitic acid, non-derivatized, GC-MS Spectrum | splash10-0002-0940000000-3ab0a8161f2ec8f04452 | Spectrum | | GC-MS | cis-Aconitic acid, non-derivatized, GC-MS Spectrum | splash10-00dj-9630000000-1dc322333cb48ef39588 | Spectrum | | GC-MS | cis-Aconitic acid, non-derivatized, GC-MS Spectrum | splash10-004i-3591000000-a58ec1ba633b098e0867 | Spectrum | | Predicted GC-MS | cis-Aconitic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-06w9-4900000000-38239492ad0e66d6533b | Spectrum | | Predicted GC-MS | cis-Aconitic acid, 3 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00di-9167000000-07ccdac5775cc3e75c95 | Spectrum | | Predicted GC-MS | cis-Aconitic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TBDMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TBDMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TBDMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TBDMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | cis-Aconitic acid, TBDMS_3_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-000i-9300000000-1f04239a9a7133f89d08 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-000f-9000000000-49f5c2a7959cfbbbe09f | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-000f-9100000000-d4857c30b96cf4786dbe | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0911000000-3197b86ab2053b968bf9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-9000000000-74977ead259f739d154b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0900000000-9f4cc240b2470f2bbb93 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0900000000-5034c256b831c6173452 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0912000000-a603cd799b70c1089ddd | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-9000000000-ce8dbec6a7b0d83b372c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0900000000-927f79de2cf5ffcb2c52 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-003r-0194000000-75d834c69fbfd0a2f7be | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-00fr-0900000000-aad30bb884e0e9a5f01c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-000i-9200000000-e90e540bcd0659005472 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-000i-9000000000-0edcc061c37b1fbb9fa1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-000l-9000000000-1e762d38e4e5cabfcb6b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-000f-9000000000-5602736cc28db9eb9085 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-000i-9400000000-c8ec5b3e604b870eab95 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00fr-0900000000-aad30bb884e0e9a5f01c | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-9200000000-e90e540bcd0659005472 | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0900000000-8e26562af9ae45eb3303 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02di-1900000000-7fec32a5e34c3b55d683 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01w0-9600000000-ee172c3013370ab0da31 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00fr-1900000000-18635ce5a20ce1b78139 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00b9-2900000000-58bb75a2a1b28dbffd73 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06rf-9300000000-19df730d01f07fcf6573 | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Citric Acid Cycle | 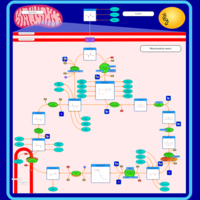 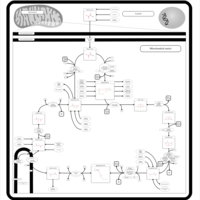 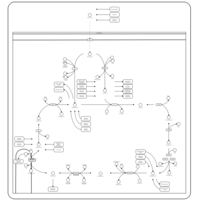 |  | | Congenital lactic acidosis | 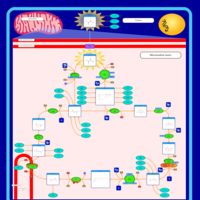 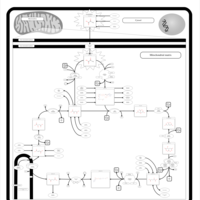 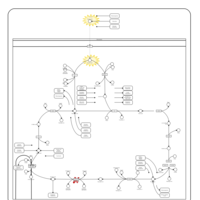 | Not Available | | Fumarase deficiency | 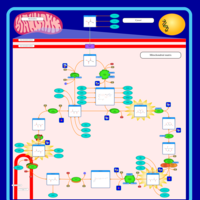 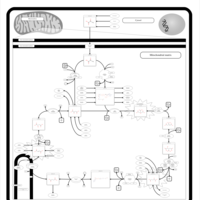 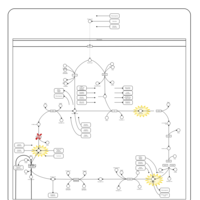 | Not Available | | Mitochondrial complex II deficiency | 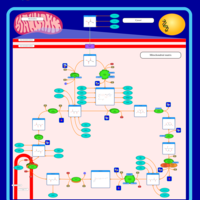 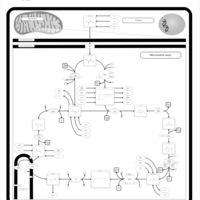 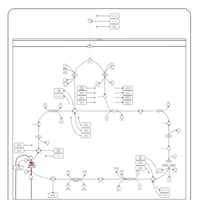 | Not Available | | 2-ketoglutarate dehydrogenase complex deficiency | 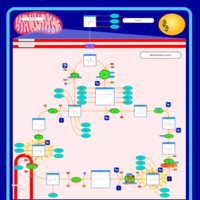 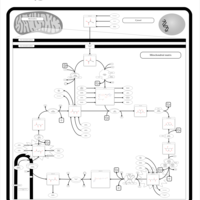 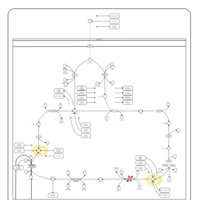 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.919 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.257 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.508 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.121 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 1.142 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.289 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000072 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB008306 |
|---|
| KNApSAcK ID | C00001177 |
|---|
| Chemspider ID | 558863 |
|---|
| KEGG Compound ID | C00417 |
|---|
| BioCyc ID | CIS-ACONITATE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Citric acid cycle |
|---|
| METLIN ID | 5130 |
|---|
| PubChem Compound | 643757 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 32805 |
|---|
| References |
|---|
| General References | Not Available |
|---|
