| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:26:00 UTC |
|---|
| Updated at | 2022-12-13 23:36:23 UTC |
|---|
| CannabisDB ID | CDB000133 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Leucine |
|---|
| Description | Leucine, abbreviated as Leu or L, l-leucine, is an essential amino acid (meaning the body cannot synthesize it and it must be obtained from the diet) that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH3+ form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain isobutyl group, making it a non-polar aliphatic amino acid. Human dietary sources are foods that contain protein, such as meats, dairy products, soy products, beans and legumes. L-Leucine is a branched-chain amino acid (BCAA). BCAAs are essential amino acids whose carbon structure is marked by a branch point. These three amino acids are critical to human life and are particularly involved in stress, energy, and muscle metabolism. BCAA supplementation as therapy, both oral and intravenous, in human health and disease holds great promise. Despite their structural similarities, the branched amino acids have different metabolic routes, with valine going solely to carbohydrates, leucine solely to fats, and isoleucine to both. The different metabolism accounts for different requirements for these essential amino acids in humans: 12 mg/kg, 14 mg/kg, and 16 mg/kg of valine, leucine, and isoleucine, respectively. Furthermore, these amino acids have different deficiency symptoms. Valine deficiency is marked by neurological defects in the brain, while isoleucine deficiency is marked by muscle tremors. Many types of inborn errors of BCAA metabolism exist and are marked by various abnormalities. The most common form is the maple syrup urine disease (PMID: 10234605 ), marked by a characteristic sweet smell of urine. Other abnormalities are associated with a wide range of symptoms, such as mental retardation, ataxia, hypoglycemia, spinal muscle atrophy, rash, vomiting, and excessive muscle movement. Most forms of BCAA metabolism errors are corrected by dietary restriction of BCAA and at least one form is correctable by supplementation with 10 mg of biotin daily. BCAA are useful because they are metabolized primarily by muscle and particularly leucine, are most essential for muscle health. BCAA and other amino acids are frequently fed intravenously (TPN) to malnourished surgical patients and, in some cases, those with severe trauma. BCAA, particularly leucine, stimulate protein synthesis, increase reutilization of amino acids in many organs and reduce protein breakdown. Furthermore, leucine can be an important source of calories, and is superior as fuel to the ubiquitous intravenous glucose (dextrose). Leucine also stimulates insulin release, which in turn stimulates protein synthesis and inhibits protein breakdown. These effects are particularly useful in athletic training. BCAA should also replace the use of steroids by weightlifters. Huntington's chorea and anorexic disorders both are characterized by low serum BCAA. These diseases, as well as forms of Parkinson's, may respond to BCAA therapy. L-Leucine can be found in most biofluids, including blood, cerebrospinal fluid (CSF), feces, and sweat, as well as throughout most human tissues. L-Leucine exists in all living species, ranging from bacteria to humans. L-leucine is also found in Cannabis plants (PMID: 6991645 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-Amino-4-methylpentanoic acid | ChEBI | | (2S)-alpha-2-Amino-4-methylvaleric acid | ChEBI | | (2S)-alpha-Leucine | ChEBI | | (S)-(+)-Leucine | ChEBI | | (S)-Leucine | ChEBI | | 2-Amino-4-methylvaleric acid | ChEBI | | L | ChEBI | | L-Leucin | ChEBI | | L-Leuzin | ChEBI | | Leu | ChEBI | | LEUCINE | ChEBI | | (2S)-2-Amino-4-methylpentanoate | Generator | | (2S)-a-2-Amino-4-methylvalerate | Generator | | (2S)-a-2-Amino-4-methylvaleric acid | Generator | | (2S)-alpha-2-Amino-4-methylvalerate | Generator | | (2S)-Α-2-amino-4-methylvalerate | Generator | | (2S)-Α-2-amino-4-methylvaleric acid | Generator | | (2S)-a-Leucine | Generator | | (2S)-Α-leucine | Generator | | 2-Amino-4-methylvalerate | Generator | | (S)-2-Amino-4-methylpentanoate | HMDB | | (S)-2-Amino-4-methylpentanoic acid | HMDB | | (S)-2-Amino-4-methylvalerate | HMDB | | (S)-2-Amino-4-methylvaleric acid | HMDB | | 4-Methyl-L-norvaline | HMDB | | L-(+)-Leucine | HMDB | | L-a-Aminoisocaproate | HMDB | | L-a-Aminoisocaproic acid | HMDB | | L-alpha-Aminoisocaproate | HMDB | | L-alpha-Aminoisocaproic acid | HMDB | | Leucine, L-isomer | HMDB | | L-Isomer leucine | HMDB | | Leucine, L isomer | HMDB |
|
|---|
| Chemical Formula | C6H13NO2 |
|---|
| Average Molecular Weight | 131.17 |
|---|
| Monoisotopic Molecular Weight | 131.0946 |
|---|
| IUPAC Name | (2S)-2-amino-4-methylpentanoic acid |
|---|
| Traditional Name | L-leucine |
|---|
| CAS Registry Number | 71000-80-1 |
|---|
| SMILES | CC(C)C[C@H](N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H13NO2/c1-4(2)3-5(7)6(8)9/h4-5H,3,7H2,1-2H3,(H,8,9)/t5-/m0/s1 |
|---|
| InChI Key | ROHFNLRQFUQHCH-YFKPBYRVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as leucine and derivatives. Leucine and derivatives are compounds containing leucine or a derivative thereof resulting from reaction of leucine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Leucine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Leucine or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Branched fatty acid
- Methyl-branched fatty acid
- Fatty acid
- Fatty acyl
- Amino acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic oxide
- Organopnictogen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Organic oxygen compound
- Amine
- Organic nitrogen compound
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 268 - 288 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 21.5 mg/mL | Not Available | | logP | -1.52 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-000l-9000000000-bf752e458f13eed8d7a2 | 2018-05-25 | View Spectrum | | GC-MS | L-Leucine, 2 TMS, GC-MS Spectrum | splash10-0pb9-0900000000-c0176b3cef05fc597576 | Spectrum | | GC-MS | L-Leucine, 2 TMS, GC-MS Spectrum | splash10-0a4i-0900000000-eb7cee37b9d78694010c | Spectrum | | GC-MS | L-Leucine, 2 TMS, GC-MS Spectrum | splash10-0a4i-0900000000-7033b5fdcd4216168462 | Spectrum | | GC-MS | L-Leucine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0900000000-cd1de48a6db61eb4455e | Spectrum | | GC-MS | L-Leucine, 2 TMS, GC-MS Spectrum | splash10-05fr-7900000000-e3b993b282ec2115b484 | Spectrum | | GC-MS | L-Leucine, non-derivatized, GC-MS Spectrum | splash10-0019-9000000000-6e468213b3429cf627bc | Spectrum | | GC-MS | L-Leucine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0900000000-ef06e48ca82519977a37 | Spectrum | | GC-MS | L-Leucine, non-derivatized, GC-MS Spectrum | splash10-0pb9-0900000000-c0176b3cef05fc597576 | Spectrum | | GC-MS | L-Leucine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0900000000-eb7cee37b9d78694010c | Spectrum | | GC-MS | L-Leucine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0900000000-7033b5fdcd4216168462 | Spectrum | | GC-MS | L-Leucine, non-derivatized, GC-MS Spectrum | splash10-0a4i-0900000000-cd1de48a6db61eb4455e | Spectrum | | GC-MS | L-Leucine, non-derivatized, GC-MS Spectrum | splash10-0udi-1391000000-5183562a14017e557e33 | Spectrum | | GC-MS | L-Leucine, non-derivatized, GC-MS Spectrum | splash10-05fr-7900000000-e3b993b282ec2115b484 | Spectrum | | Predicted GC-MS | L-Leucine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9000000000-70cdb961a0819be4318a | Spectrum | | Predicted GC-MS | L-Leucine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-000i-9100000000-991398731b9d8305622c | Spectrum | | Predicted GC-MS | L-Leucine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Leucine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Leucine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Leucine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Leucine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-000i-9100000000-05b5a7a191a32803595e | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0006-9000000000-5302e9c96e75e06e9705 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0006-9000000000-9e70778b46864cd22996 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80) , Positive | splash10-0019-9000000000-6e468213b3429cf627bc | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-4f403c61aaa8a103c049 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-d85fcc558423654f45fd | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-931937d3bdd49b3ae623 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0930000000-aa1256c9224fecacf025 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-0a72425a86f804d3a16b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-4ed8cafebf4ee4e82e6a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-3ac62b780abd90dd28b1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-eb75eb03a58512ea36df | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-001i-0900000000-c0f28e4ebdef67c5b509 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-001i-0900000000-0751a9e803a2e7715d1a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-001i-5900000000-8040fc883917239630a7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-016s-9000000000-a7f9e8d43e66dbd02b6f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-001r-7900000000-da119558c426d9c9d3a1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-000i-9000000000-ae8365105ae2a18e0c10 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-000f-9000000000-228ee614be6648b37433 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-0006-9000000000-d1f3e047af455e156a1e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-0006-9000000000-409dfbdb20719c3afda5 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - CE-ESI-TOF (CE-system connected to 6210 Time-of-Flight MS, Agilent) , Positive | splash10-001i-0900000000-720554d58264a9cfdb67 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001i-0900000000-90fab591d1f6d6318002 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-001i-0900000000-ed843252a559d532dc5e | 2012-08-31 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0019-9500000000-b9f1203176ce0ca7c11c | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Valine, Leucine and Isoleucine Degradation |  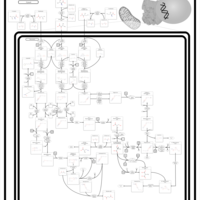 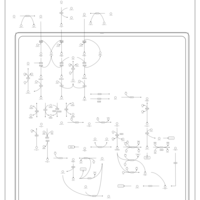 |  | | Transcription/Translation | Not Available | Not Available | | Beta-Ketothiolase Deficiency |  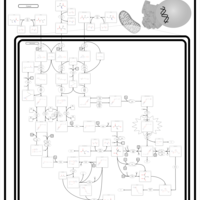 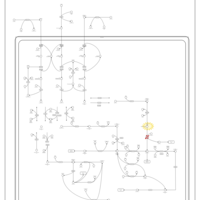 | Not Available | | 2-Methyl-3-Hydroxybutryl CoA Dehydrogenase Deficiency |  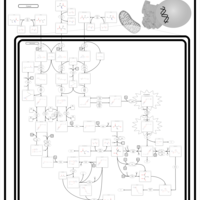 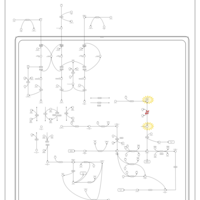 | Not Available | | Propionic Acidemia | 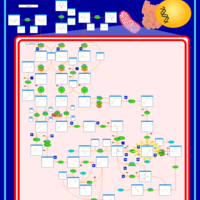 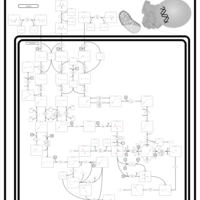 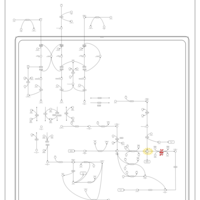 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Branched-chain-amino-acid aminotransferase, cytosolic | BCAT1 | 12p12.1 | P54687 | details | | Branched-chain-amino-acid aminotransferase, mitochondrial | BCAT2 | 19q13 | O15382 | details | | Probable leucine--tRNA ligase, mitochondrial | LARS2 | 3p21.3 | Q15031 | details | | Leucine--tRNA ligase, cytoplasmic | LARS | 5q32 | Q9P2J5 | details | | Leucine carboxyl methyltransferase 2 | LCMT2 | 15q15.3 | O60294 | details | | Leucine carboxyl methyltransferase 1 | LCMT1 | 16p12.1 | Q9UIC8 | details |
|
|---|
| Transporters | |
| Large neutral amino acids transporter small subunit 2 | SLC7A8 | 14q11.2 | Q9UHI5 | details |
|
|---|
| Metal Bindings | Not Available |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.303 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.159 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.207 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.184 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.0575 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.165 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000687 |
|---|
| DrugBank ID | DB00149 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000899 |
|---|
| KNApSAcK ID | C00001377 |
|---|
| Chemspider ID | 5880 |
|---|
| KEGG Compound ID | C00123 |
|---|
| BioCyc ID | LEU |
|---|
| BiGG ID | 33942 |
|---|
| Wikipedia Link | Leucine |
|---|
| METLIN ID | 24 |
|---|
| PubChem Compound | 6106 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15603 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Podebrad F, Heil M, Reichert S, Mosandl A, Sewell AC, Bohles H: 4,5-dimethyl-3-hydroxy-2[5H]-furanone (sotolone)--the odour of maple syrup urine disease. J Inherit Metab Dis. 1999 Apr;22(2):107-14. doi: 10.1023/a:1005433516026. [PubMed:10234605 ]
|
|---|
