| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:42:49 UTC |
|---|
| Updated at | 2020-12-07 19:11:05 UTC |
|---|
| CannabisDB ID | CDB004846 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Arginine |
|---|
| Description | Arginine or l-arginine, abbreviated Arg or R, is an essential amino acid that is physiologically active in the L-form. In mammals, arginine is formally classified as a semi-essential or conditionally essential amino acid, depending on the developmental stage and health status of the individual. Pre-term infants are unable to effectively synthesize arginine, making it nutritionally essential for them. Adults, however, synthesize arginine from glutamate via citrulline. Arginine is a basic amino acid as its side chain contains a positively charged guanidinium group, which is highly polar, at the end of a hydrophobic aliphatic hydrocarbon chain. With a pKa of 12.48, the guanidinium group is positively charged in neutral, acidic, and even most basic environments. Because of the conjugation between the double bond and the nitrogen lone pairs, the positive charge is delocalized. This group is able to form multiple H-bonds. L-Arginine is an amino acid that has numerous functions in the body. It helps dispose of ammonia, is a precursor of nitric oxide, creatine, L-glutamate, and L-proline, and it can be converted into glucose and glycogen if needed. In large doses, L-arginine also stimulates the release of the hormones, growth hormone and prolactin. Arginine is a known inducer of mTOR (mammalian target of rapamycin) and is responsible for inducing protein synthesis through the mTOR pathway. mTOR inhibition by rapamycin partially reduces arginine-induced protein synthesis (PMID: 20841502 ). Catabolic disease states such as sepsis, injury, and cancer cause an increase in arginine utilization, which can exceed normal body production, leading to arginine depletion. Arginine also activates AMP kinase (AMPK) which then stimulates skeletal muscle fatty acid oxidation and muscle glucose uptake, thereby increasing insulin secretion by pancreatic beta-cells (PMID: 21311355 ). Arginine is found in plant and animal proteins, such as dairy products, meat, poultry, fish, and nuts. While many studies suggested that plant based protein-rich foods like soy protein, which contain less arginine, reduced cardiovascular risk compared to arginine rich foods like animal proteins, other studies have found little or null effects (PMID: 20042191 ). Arginine is one of the amino acids that are known in cannabis plant (PMID: 6991645 ). L-Arginine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-Amino-5-(carbamimidamido)pentanoic acid | ChEBI | | (2S)-2-Amino-5-guanidinopentanoic acid | ChEBI | | (S)-2-Amino-5-guanidinopentanoic acid | ChEBI | | (S)-2-Amino-5-guanidinovaleric acid | ChEBI | | Arg | ChEBI | | Arginine | ChEBI | | L-(+)-Arginine | ChEBI | | L-Arg | ChEBI | | L-Arginin | ChEBI | | R | ChEBI | | (2S)-2-Amino-5-(carbamimidamido)pentanoate | Generator | | (2S)-2-Amino-5-guanidinopentanoate | Generator | | (S)-2-Amino-5-guanidinopentanoate | Generator | | (S)-2-Amino-5-guanidinovalerate | Generator | | (S)-2-Amino-5-[(aminoiminomethyl)amino]-pentanoate | HMDB | | (S)-2-Amino-5-[(aminoiminomethyl)amino]-pentanoic acid | HMDB | | (S)-2-Amino-5-[(aminoiminomethyl)amino]pentanoate | HMDB | | (S)-2-Amino-5-[(aminoiminomethyl)amino]pentanoic acid | HMDB | | 2-Amino-5-guanidinovalerate | HMDB | | 2-Amino-5-guanidinovaleric acid | HMDB | | 5-[(Aminoiminomethyl)amino]-L-norvaline | HMDB | | L-a-Amino-D-guanidinovalerate | HMDB | | L-a-Amino-D-guanidinovaleric acid | HMDB | | L-alpha-Amino-delta-guanidinovalerate | HMDB | | L-alpha-Amino-delta-guanidinovaleric acid | HMDB | | N5-(Aminoiminomethyl)-L-ornithine | HMDB | | DL-Arginine acetate, monohydrate | HMDB | | L-Isomer arginine | HMDB | | Monohydrate DL-arginine acetate | HMDB | | L Arginine | HMDB | | Arginine, L isomer | HMDB | | Arginine, L-isomer | HMDB | | Hydrochloride, arginine | HMDB | | Arginine hydrochloride | HMDB | | DL Arginine acetate, monohydrate | HMDB |
|
|---|
| Chemical Formula | C6H14N4O2 |
|---|
| Average Molecular Weight | 174.2 |
|---|
| Monoisotopic Molecular Weight | 174.1117 |
|---|
| IUPAC Name | (2S)-2-amino-5-carbamimidamidopentanoic acid |
|---|
| Traditional Name | L-arginine |
|---|
| CAS Registry Number | 74-79-3 |
|---|
| SMILES | N[C@@H](CCCNC(N)=N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H14N4O2/c7-4(5(11)12)2-1-3-10-6(8)9/h4H,1-3,7H2,(H,11,12)(H4,8,9,10)/t4-/m0/s1 |
|---|
| InChI Key | ODKSFYDXXFIFQN-BYPYZUCNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Fatty acid
- Guanidine
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Carboximidamide
- Propargyl-type 1,3-dipolar organic compound
- Organic 1,3-dipolar compound
- Amine
- Hydrocarbon derivative
- Organic oxide
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Primary aliphatic amine
- Organic oxygen compound
- Organic nitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 222 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 182 mg/mL at 25 °C | Human Metabolome Project | | logP | -4.20 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | L-Arginine, 3 TMS, GC-MS Spectrum | splash10-0a4i-1910000000-0191c1a63652c493660b | Spectrum | | GC-MS | L-Arginine, 3 TMS, GC-MS Spectrum | splash10-00di-9810000000-eb6eb73302b678cf0a24 | Spectrum | | GC-MS | L-Arginine, 2 TMS, GC-MS Spectrum | splash10-00di-9700000000-e47b41cff0e873f53932 | Spectrum | | GC-MS | L-Arginine, 3 TMS, GC-MS Spectrum | splash10-0a4i-1920000000-8ae5af11398835d26bed | Spectrum | | GC-MS | L-Arginine, non-derivatized, GC-MS Spectrum | splash10-0a4i-1910000000-0191c1a63652c493660b | Spectrum | | GC-MS | L-Arginine, non-derivatized, GC-MS Spectrum | splash10-00di-9810000000-eb6eb73302b678cf0a24 | Spectrum | | GC-MS | L-Arginine, non-derivatized, GC-MS Spectrum | splash10-00di-9700000000-e47b41cff0e873f53932 | Spectrum | | GC-MS | L-Arginine, non-derivatized, GC-MS Spectrum | splash10-0a4i-1920000000-8ae5af11398835d26bed | Spectrum | | GC-MS | L-Arginine, non-derivatized, GC-MS Spectrum | splash10-0udi-0900000000-afc3ca93f8fbf54ec9f4 | Spectrum | | Predicted GC-MS | L-Arginine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00dl-9100000000-60db59de76b9a9b5bfc8 | Spectrum | | Predicted GC-MS | L-Arginine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-004i-8900000000-f47c4f9480612fdbc962 | Spectrum | | Predicted GC-MS | L-Arginine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, TMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, TBDMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, TBDMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Arginine, TBDMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0fk9-0946231100-3d57d2304dcb33feab3f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-000i-0900000000-5b6dd6fb263ea09289fc | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0900000000-dfe35b3438d19320d8cb | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0900000000-f666ab7e5354bce67a2e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03kd-0977452210-046c4b70bd0ec351c41d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-014i-9000000000-2f51e43e530976d63633 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0900000000-b9ebb7ebccee1a313888 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-0900000000-5379b6fb6ea2313101f9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-001i-0900000000-835751d54af24bd337e7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-001i-0900000000-2d2b5fd7617ccb227bb1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-001i-0900000000-dfe35b3438d19320d8cb | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-00di-0900000000-f666ab7e5354bce67a2e | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-001i-0900000000-b9ebb7ebccee1a313888 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-00di-0900000000-5379b6fb6ea2313101f9 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-001i-0900000000-835751d54af24bd337e7 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-001i-0900000000-2d2b5fd7617ccb227bb1 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-001i-0900000000-8e090b4f0a569ec011b3 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900000000-647e6324d4041e9d5cc1 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-d866a7f7b1bf575941ee | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-001i-1900000000-c3a3ef3d4f3627781019 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00di-3900000000-8c82418f7b35a97fb9b3 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00di-9000000000-19b62da79866318c52dd | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00di-9000000000-64b046d21bdcbb8d5923 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-004i-0900000000-e07b937b6867d1f62293 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4i-0900000000-87ab853583aab2973cfb | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Urea Cycle | 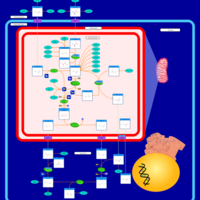 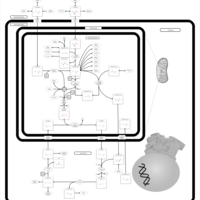 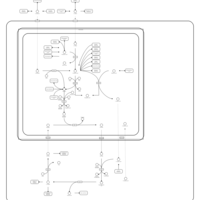 |  | | Transcription/Translation | Not Available | Not Available | | Arginine and Proline Metabolism | 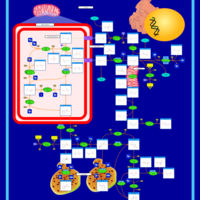 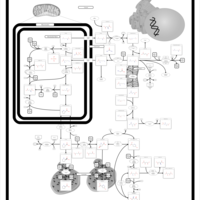 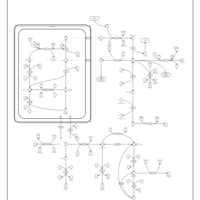 |  | | Glycine and Serine Metabolism | 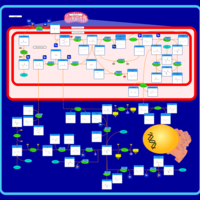 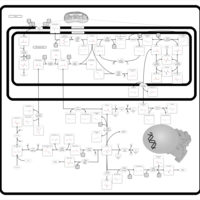 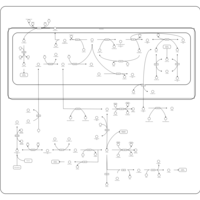 |  | | Prolidase Deficiency (PD) | 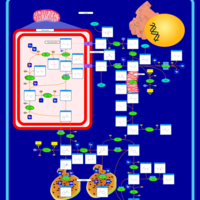 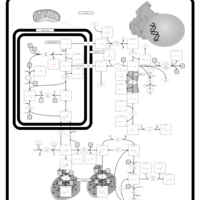 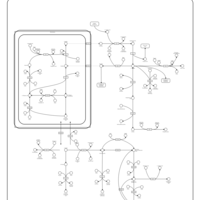 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000517 |
|---|
| DrugBank ID | DB00125 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB002257 |
|---|
| KNApSAcK ID | C00001340 |
|---|
| Chemspider ID | 6082 |
|---|
| KEGG Compound ID | C00062 |
|---|
| BioCyc ID | ARG |
|---|
| BiGG ID | 33707 |
|---|
| Wikipedia Link | L-arginine |
|---|
| METLIN ID | 5502 |
|---|
| PubChem Compound | 6322 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16467 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Bauchart-Thevret C, Cui L, Wu G, Burrin DG: Arginine-induced stimulation of protein synthesis and survival in IPEC-J2 cells is mediated by mTOR but not nitric oxide. Am J Physiol Endocrinol Metab. 2010 Dec;299(6):E899-909. doi: 10.1152/ajpendo.00068.2010. Epub 2010 Sep 14. [PubMed:20841502 ]
- Linden KC, Wadley GD, Garnham AP, McConell GK: Effect of l-arginine infusion on glucose disposal during exercise in humans. Med Sci Sports Exerc. 2011 Sep;43(9):1626-34. doi: 10.1249/MSS.0b013e318212a317. [PubMed:21311355 ]
- Vega-Lopez S, Matthan NR, Ausman LM, Harding SV, Rideout TC, Ai M, Otokozawa S, Freed A, Kuvin JT, Jones PJ, Schaefer EJ, Lichtenstein AH: Altering dietary lysine:arginine ratio has little effect on cardiovascular risk factors and vascular reactivity in moderately hypercholesterolemic adults. Atherosclerosis. 2010 Jun;210(2):555-62. doi: 10.1016/j.atherosclerosis.2009.12.002. Epub 2009 Dec 5. [PubMed:20042191 ]
|
|---|

